(Notice): Undefined variable: solution_of_interest [APP/View/Products/view.ctp, line 755]Code Context $errorType = self::getErrorType($code);
self::logToDatabase( $errorType, $description, $file, $line);
return parent::handleError( $errorType, $description, $file, $line, $context);
$viewFile = '/var/www/dev.diagenode.com/app/View/Products/view.ctp'
$dataForView = array(
'language' => 'en',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'meta_title' => 'ATAC-seq kit for open chromatin assessment C01080001 | Diagenode ',
'product' => array(
'Product' => array(
'id' => '3194',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<div class="row">
<div class="small-12 medium-8 large-8 columns">
<p><a class="tiny details button" href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf">Download the Manual</a></p>
<p><strong>ATAC-seq</strong>, Assay for<span> </span><strong>T</strong>ransposase-<strong>A</strong>ccessible<span> </span><strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the<span> </span><strong>transposase Tn5</strong><span> </span>which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
</div>
<div class="small-12 medium-4 large-4 columns"><center>
<script>// <![CDATA[
var date = new Date(); var heure = date.getHours(); var jour = date.getDay(); var semaine = Math.floor(date.getDate() / 7) + 1; if (jour === 2 && ( (heure >= 9 && heure < 9.5) || (heure >= 18 && heure < 18.5) )) { document.write('<a href="https://us02web.zoom.us/j/85467619762"><img src="https://www.diagenode.com/img/epicafe-ON.gif"></a>'); } else { document.write('<a href="https://go.diagenode.com/l/928883/2023-04-26/3kq1v"><img src="https://www.diagenode.com/img/epicafe-OFF.png"></a>'); }
// ]]></script>
</center></div>
</div>
<p>Diagenode’s<span> </span><b>ATAC-</b><b>seq</b><b><span> </span>kit<span> </span></b>is based on a highly validated protocol optimized for<span> </span><b>50,000<span> </span></b><b>cells</b><b><span> </span>per<span> </span></b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The <a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">primer indexes for multiplexing</a> are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell<span> </span></b><b>requirement</b><b>:<span> </span></b><b>50,000<span> </span></b><b>cells /<span> </span></b><b>rxn</b></li>
<li><b>Robust protocol<span> </span></b>with<span> </span><b>high reproducibility<span> </span></b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong><span> </span>and<span> </span><b>efficient DNA capture<span> </span></b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids<span> </span></b><b>over-amplification</b></li>
<li>Allows adaptation/flexibility for<span> </span><b>more challenging samples<span> </span></b>to succeed with library prep.</li>
<li>Gives<span> </span><strong>early indication</strong><span> </span>if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p>Looking for ATAC-seq on tissue? Please, go to: <a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
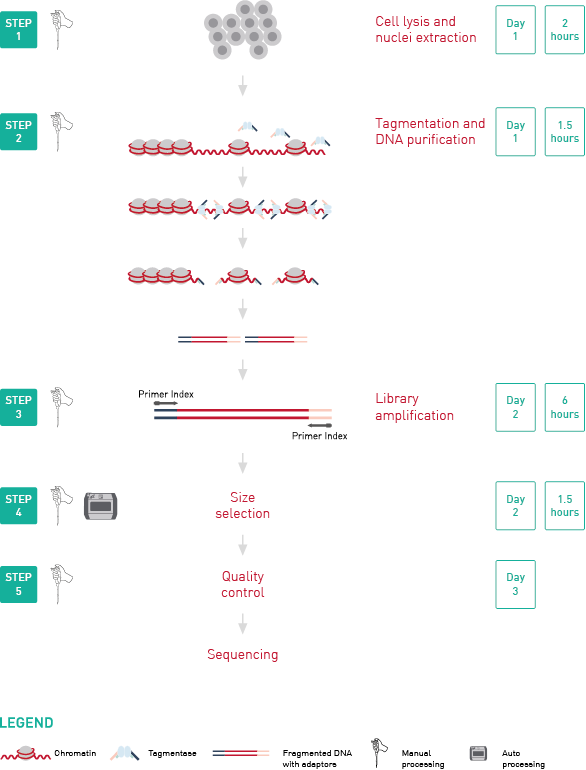
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
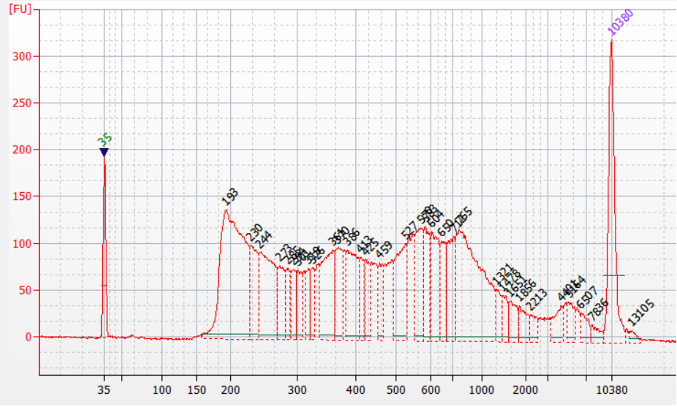
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
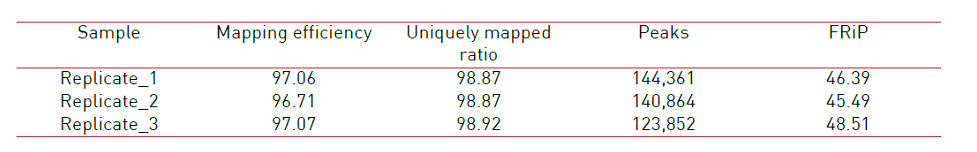
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
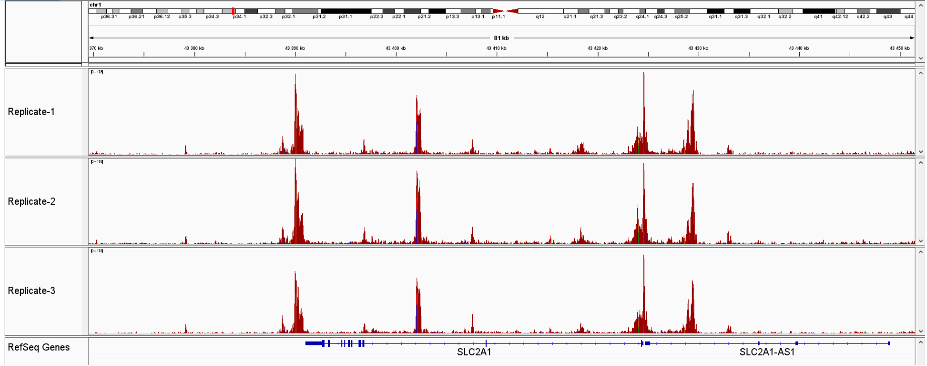
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
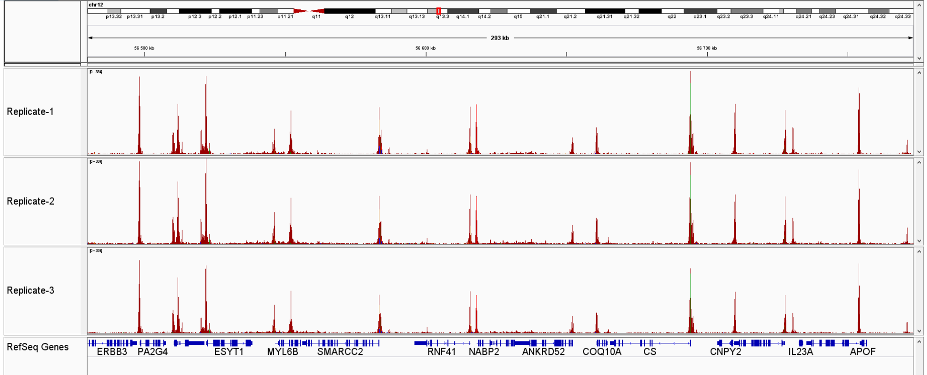
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
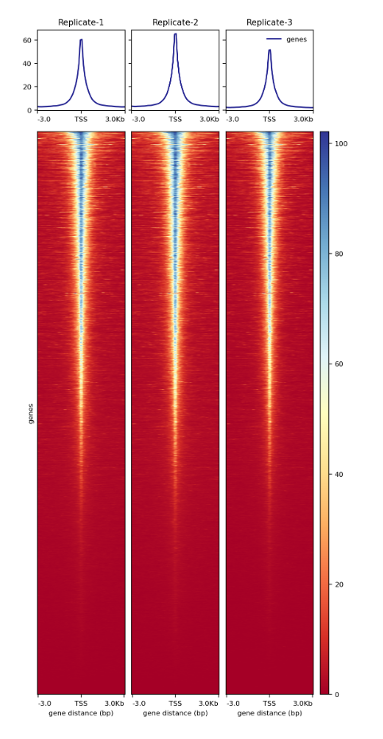
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack:<span> </span><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"><span> </span>0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"><span> </span>(Tn5 transposase)<span> </span></a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"><span> </span>Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '24 rxns',
'catalog_number' => 'C01080002',
'old_catalog_number' => '',
'sf_code' => 'C01080002-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '1740',
'price_USD' => '1745',
'price_GBP' => '1485',
'price_JPY' => '313200',
'price_CNY' => '',
'price_AUD' => '4362',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-24rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment C01080001 | Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2025-04-17 10:29:58',
'created' => '2021-07-01 15:46:58',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => null,
'name' => null,
'description' => null,
'clonality' => null,
'isotype' => null,
'lot' => null,
'concentration' => null,
'reactivity' => null,
'type' => null,
'purity' => null,
'classification' => null,
'application_table' => null,
'storage_conditions' => null,
'storage_buffer' => null,
'precautions' => null,
'uniprot_acc' => null,
'slug' => null,
'meta_keywords' => null,
'meta_description' => null,
'modified' => null,
'created' => null,
'select_label' => null
),
'Slave' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Group' => array(
'Group' => array(
[maximum depth reached]
),
'Master' => array(
[maximum depth reached]
),
'Product' => array(
[maximum depth reached]
)
),
'Related' => array(),
'Application' => array(),
'Category' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Document' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
),
(int) 8 => array(
[maximum depth reached]
),
(int) 9 => array(
[maximum depth reached]
),
(int) 10 => array(
[maximum depth reached]
),
(int) 11 => array(
[maximum depth reached]
),
(int) 12 => array(
[maximum depth reached]
),
(int) 13 => array(
[maximum depth reached]
),
(int) 14 => array(
[maximum depth reached]
),
(int) 15 => array(
[maximum depth reached]
),
(int) 16 => array(
[maximum depth reached]
),
(int) 17 => array(
[maximum depth reached]
),
(int) 18 => array(
[maximum depth reached]
),
(int) 19 => array(
[maximum depth reached]
),
(int) 20 => array(
[maximum depth reached]
),
(int) 21 => array(
[maximum depth reached]
),
(int) 22 => array(
[maximum depth reached]
),
(int) 23 => array(
[maximum depth reached]
),
(int) 24 => array(
[maximum depth reached]
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
)
)
),
'meta_canonical' => 'https://dev.diagenode.com/en/p/atac-seq-kit-24rxns',
'fromEmail' => 'dev.diagenode.com'
)
$language = 'en'
$meta_keywords = ''
$meta_description = 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility'
$meta_title = 'ATAC-seq kit for open chromatin assessment C01080001 | Diagenode '
$product = array(
'Product' => array(
'id' => '3194',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<div class="row">
<div class="small-12 medium-8 large-8 columns">
<p><a class="tiny details button" href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf">Download the Manual</a></p>
<p><strong>ATAC-seq</strong>, Assay for<span> </span><strong>T</strong>ransposase-<strong>A</strong>ccessible<span> </span><strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the<span> </span><strong>transposase Tn5</strong><span> </span>which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
</div>
<div class="small-12 medium-4 large-4 columns"><center>
<script>// <![CDATA[
var date = new Date(); var heure = date.getHours(); var jour = date.getDay(); var semaine = Math.floor(date.getDate() / 7) + 1; if (jour === 2 && ( (heure >= 9 && heure < 9.5) || (heure >= 18 && heure < 18.5) )) { document.write('<a href="https://us02web.zoom.us/j/85467619762"><img src="https://www.diagenode.com/img/epicafe-ON.gif"></a>'); } else { document.write('<a href="https://go.diagenode.com/l/928883/2023-04-26/3kq1v"><img src="https://www.diagenode.com/img/epicafe-OFF.png"></a>'); }
// ]]></script>
</center></div>
</div>
<p>Diagenode’s<span> </span><b>ATAC-</b><b>seq</b><b><span> </span>kit<span> </span></b>is based on a highly validated protocol optimized for<span> </span><b>50,000<span> </span></b><b>cells</b><b><span> </span>per<span> </span></b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The <a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">primer indexes for multiplexing</a> are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell<span> </span></b><b>requirement</b><b>:<span> </span></b><b>50,000<span> </span></b><b>cells /<span> </span></b><b>rxn</b></li>
<li><b>Robust protocol<span> </span></b>with<span> </span><b>high reproducibility<span> </span></b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong><span> </span>and<span> </span><b>efficient DNA capture<span> </span></b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids<span> </span></b><b>over-amplification</b></li>
<li>Allows adaptation/flexibility for<span> </span><b>more challenging samples<span> </span></b>to succeed with library prep.</li>
<li>Gives<span> </span><strong>early indication</strong><span> </span>if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p>Looking for ATAC-seq on tissue? Please, go to: <a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack:<span> </span><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"><span> </span>0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"><span> </span>(Tn5 transposase)<span> </span></a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"><span> </span>Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '24 rxns',
'catalog_number' => 'C01080002',
'old_catalog_number' => '',
'sf_code' => 'C01080002-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '1740',
'price_USD' => '1745',
'price_GBP' => '1485',
'price_JPY' => '313200',
'price_CNY' => '',
'price_AUD' => '4362',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-24rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment C01080001 | Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2025-04-17 10:29:58',
'created' => '2021-07-01 15:46:58',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => null,
'name' => null,
'description' => null,
'clonality' => null,
'isotype' => null,
'lot' => null,
'concentration' => null,
'reactivity' => null,
'type' => null,
'purity' => null,
'classification' => null,
'application_table' => null,
'storage_conditions' => null,
'storage_buffer' => null,
'precautions' => null,
'uniprot_acc' => null,
'slug' => null,
'meta_keywords' => null,
'meta_description' => null,
'modified' => null,
'created' => null,
'select_label' => null
),
'Slave' => array(
(int) 0 => array(
'id' => '349',
'name' => 'C01080002',
'product_id' => '3194',
'modified' => '2021-07-01 15:47:50',
'created' => '2021-07-01 15:47:50'
)
),
'Group' => array(
'Group' => array(
'id' => '349',
'name' => 'C01080002',
'product_id' => '3194',
'modified' => '2021-07-01 15:47:50',
'created' => '2021-07-01 15:47:50'
),
'Master' => array(
'id' => '3194',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<div class="row">
<div class="small-12 medium-8 large-8 columns">
<p><a class="tiny details button" href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf">Download the Manual</a></p>
<p><strong>ATAC-seq</strong>, Assay for<span> </span><strong>T</strong>ransposase-<strong>A</strong>ccessible<span> </span><strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the<span> </span><strong>transposase Tn5</strong><span> </span>which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
</div>
<div class="small-12 medium-4 large-4 columns"><center>
<script>// <![CDATA[
var date = new Date(); var heure = date.getHours(); var jour = date.getDay(); var semaine = Math.floor(date.getDate() / 7) + 1; if (jour === 2 && ( (heure >= 9 && heure < 9.5) || (heure >= 18 && heure < 18.5) )) { document.write('<a href="https://us02web.zoom.us/j/85467619762"><img src="https://www.diagenode.com/img/epicafe-ON.gif"></a>'); } else { document.write('<a href="https://go.diagenode.com/l/928883/2023-04-26/3kq1v"><img src="https://www.diagenode.com/img/epicafe-OFF.png"></a>'); }
// ]]></script>
</center></div>
</div>
<p>Diagenode’s<span> </span><b>ATAC-</b><b>seq</b><b><span> </span>kit<span> </span></b>is based on a highly validated protocol optimized for<span> </span><b>50,000<span> </span></b><b>cells</b><b><span> </span>per<span> </span></b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The <a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">primer indexes for multiplexing</a> are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell<span> </span></b><b>requirement</b><b>:<span> </span></b><b>50,000<span> </span></b><b>cells /<span> </span></b><b>rxn</b></li>
<li><b>Robust protocol<span> </span></b>with<span> </span><b>high reproducibility<span> </span></b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong><span> </span>and<span> </span><b>efficient DNA capture<span> </span></b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids<span> </span></b><b>over-amplification</b></li>
<li>Allows adaptation/flexibility for<span> </span><b>more challenging samples<span> </span></b>to succeed with library prep.</li>
<li>Gives<span> </span><strong>early indication</strong><span> </span>if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p>Looking for ATAC-seq on tissue? Please, go to: <a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack:<span> </span><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"><span> </span>0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"><span> </span>(Tn5 transposase)<span> </span></a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"><span> </span>Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '24 rxns',
'catalog_number' => 'C01080002',
'old_catalog_number' => '',
'sf_code' => 'C01080002-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '1740',
'price_USD' => '1745',
'price_GBP' => '1485',
'price_JPY' => '313200',
'price_CNY' => '',
'price_AUD' => '4362',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-24rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment C01080001 | Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2025-04-17 10:29:58',
'created' => '2021-07-01 15:46:58'
),
'Product' => array(
(int) 0 => array(
[maximum depth reached]
)
)
),
'Related' => array(),
'Application' => array(),
'Category' => array(
(int) 0 => array(
'id' => '13',
'position' => '2',
'parent_id' => null,
'name' => 'Chromatin studies',
'description' => '<div class="row">
<div class="small-12 medium-8 large-8 columns"><br />
<p>Chromatin structure plays a key role in regulating gene expression by allowing DNA accessibility to transcriptional machinery and transcription factors. The packaging of DNA into nucleosomes forms a closed structure that is not highly accessible to transcriptional elements whereas the open nucleosome structure allows DNA to be accessible. Diagenode offers a number of solutions to help you analyze chromatin and the role of transcriptional machinery including ChIP kits, ChIPmentation kits, antibodies, pA-Tn5 and ATAC-seq kits.</p>
</div>
<div class="small-12 medium-4 large-4 columns"><center>
<script>// <![CDATA[
var date = new Date(); var heure = date.getHours(); var jour = date.getDay(); var semaine = Math.floor(date.getDate() / 7) + 1; if (jour === 2 && ( (heure >= 9 && heure < 9.5) || (heure >= 18 && heure < 18.5) )) { document.write('<a href="https://us02web.zoom.us/j/85467619762"><img src="https://www.diagenode.com/img/epicafe-ON.gif"></a>'); } else { document.write('<a href="https://go.diagenode.com/l/928883/2023-04-26/3kq1v"><img src="https://www.diagenode.com/img/epicafe-OFF.png"></a>'); }
// ]]></script>
</center></div>
</div>
<p><a href="https://www.diagenode.com/en/p/uchipmentation-for-histones-24-rxns"><img src="https://www.diagenode.com/img/banners/b-microchip-category.png" /></a></p>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<div id="portal" class="main-portal">
<div class="portal-inner"><nav class="portal-nav">
<ul class="tips-menu">
<li><a href="#workflow" class="tips portal button" style="background: #13b29c; color: #f3fbfa;">Chromatin immunoprecipitation</a></li>
<li><a href="https://www.diagenode.com/en/categories/chromatin-ip-chipmentation" class="tips portal button">ChIPmentation</a></li>
<li><a href="https://www.diagenode.com/en/categories/antibodies
" class="tips portal button">Antibodies</a></li>
<li><a href="https://www.diagenode.com/en/categories/cutandtag" class="tips portal button">pA-Tn5</a></li>
<li><a href="https://www.diagenode.com/en/categories/atac-seq" class="tips portal button">ATAC-seq</a></li>
<li><a href="https://www.diagenode.com/en/categories/cutandtag" class="tips portal button">CUT&Tag</a></li>
</ul>
</nav></div>
</div>
<p>Chromatin immunoprecipitation (ChIP) determines the location of DNA binding sites on the genome for a protein of interest, giving insights into gene expression regulation. ChIP involves the selective enrichment of a chromatin fraction containing a specific antigen. Antibodies that recognize a protein or protein modification are used to determine the relative abundance of that antigen at a specific locus or loci. ChIP can be used to compare enrichment of proteins, map protein modifications, or quantify a protein modification during a time course.</p>
<span class="anchor" id="workflow"></span>
<h4><span style="font-weight: 400;">The ChIP workflow</span></h4>
<ul class="accordion" data-accordion="">
<li class="accordion-navigation"><img src="https://www.diagenode.com/img/categories/kits_chromatin_function/website-chip-workflow.jpg" />
<div id="chip_workflow" class="content">
<div class="row">
<table>
<tbody>
<tr>
<td width="50%">
<h3 class="text-center">Step by step workflow</h3>
</td>
<td width="50%">
<h3 class="text-center">Optimal solution from Diagenode</h3>
</td>
</tr>
<tr>
<td colspan="2"></td>
</tr>
<tr valign="top">
<td>
<p class="lead text-center"><strong>1.</strong><span> </span>Crosslink to bind proteins to DNA</p>
<center><img src="https://www.diagenode.com/img/chip/step-by-step-1-workflow.png" width="192" height="24" /></center></td>
<td>
<ul class="arrow">
<li><a href="https://www.diagenode.com/p/chip-cross-link-gold-600-ul">ChIP Cross-link Gold</a></li>
</ul>
</td>
</tr>
<tr>
<td colspan="2"></td>
</tr>
<tr valign="top">
<td>
<p class="lead text-center"><strong>2.</strong><span> </span>Shear DNA</p>
<center><img src="https://www.diagenode.com/img/chip/step-by-step-2-workflow.png" width="194" height="80" /></center></td>
<td>
<ul class="arrow">
<li><a href="https://www.diagenode.com/p/bioruptor-pico-sonication-device#">Bioruptor<sup>®</sup><span> </span>Pico Sonication device</a></li>
<li><a href="https://www.diagenode.com/categories/chromatin-shearing">Shearing optimization reagent</a></li>
</ul>
</td>
</tr>
<tr>
<td colspan="2"></td>
</tr>
<tr valign="top">
<td>
<p class="lead text-center"><strong>3.</strong><span> </span>Immunoprecipitate with specific antibody</p>
<center><img src="https://www.diagenode.com/img/chip/step-by-step-3-workflow.png" width="47" height="51" /></center></td>
<td>
<ul class="arrow">
<li><a href="https://www.diagenode.com/categories/chromatin-immunoprecipitation">ChIP-seq and ChIP-qPCR kits</a><span> </span>for transcription factors, histones, low inputs, plants</li>
<li><a href="https://www.diagenode.com/categories/chip-grade-antibodies">ChIP</a><span> </span>and<span> </span><a href="https://www.diagenode.com/categories/chip-seq-grade-antibodies">ChIP-seq grade</a><span> </span>antibodies</li>
<li><a href="https://www.diagenode.com/categories/ip-star">Automation available</a></li>
</ul>
</td>
</tr>
<tr>
<td colspan="2"></td>
</tr>
<tr valign="top">
<td>
<p class="lead text-center"><strong>4.</strong><span> </span>Reverse crosslinks and purify</p>
<center><img src="https://www.diagenode.com/img/chip/step-by-step-4-workflow.png" width="194" height="80" /></center></td>
<td>
<ul class="arrow">
<li>Diagenode's ChIP kits (contain optimal purification modules)</li>
</ul>
</td>
</tr>
<tr>
<td colspan="2"></td>
</tr>
<tr valign="top">
<td>
<p class="lead text-center"><strong>5.</strong><span> </span><a href="https://www.diagenode.com/en/p/ideal-chip-qpcr-kit">ChIP-qPCR</a><span> </span>or ChIP-seq library preparation</p>
</td>
<td>
<ul class="arrow">
<li>ChIP-seq:</li>
</ul>
<ul style="list-style-type: square;">
<li style="margin-left: 40px; font-size: 1.09rem;"><a href="https://www.diagenode.com/p/microplex-library-preparation-kit-v2-x48-12-indices-48-rxns">MicroPlex Library Preparation for 50 pg - 5 ng</a></li>
<li style="margin-left: 40px; font-size: 1.09rem;"><a href="https://www.diagenode.com/p/ideal-library-preparation-kit-x24-incl-index-primer-set-1-24-rxns">iDeal Library Preparation for > 5 ng</a></li>
</ul>
<ul class="arrow">
<li>ChIP qPCR</li>
</ul>
</td>
</tr>
</tbody>
</table>
</div>
</div>
</li>
</ul>
<p></p>
<h4><span style="font-weight: 400;">Products for chromatin study</span></h4>
<p><span style="font-weight: 400;"></span></p>
<p style="padding-left: 30px;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/crosslinking.png" alt="" width="35" height="35" /> Crosslinking<br /></b></span></strong><span style="font-weight: 400;">Efficient solution for protein-protein fixation.</span><strong><span style="font-weight: 400;"><b><span style="font-weight: 400;"> <a href="../p/chip-cross-link-gold-600-ul
">Read more</a></span></b><br /></span></strong></p>
<p style="padding-left: 30px;"><strong><img src="https://www.diagenode.com/img/applications/chromatin-shearing.png" alt="" width="35" height="35" /> Chromatin shearing<br /></strong><strong><span style="font-weight: 400;">Perfectly sheared chromatin is critical for ChIP success.<span> </span><br /></span></strong><strong><a href="../categories/chromatin-shearing"><span style="font-weight: 400;">Read more</span></a><span style="font-weight: 400;"><span> </span>about solutions for successful chromatin preparation.<span> </span><br /></span></strong><strong><span style="font-weight: 400;"><a href="../categories/bioruptor-shearing-device">Read more</a><span> </span>about<span> </span></span><span style="font-weight: 400;">Bioruptor<span> </span></span><span style="font-weight: 400;">sonication.</span></strong></p>
<p style="padding-left: 30px;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/immunoprecipitation.png" width="35" height="35" caption="false" /> Chromatin immunoprecipitation<br /></b></span></strong><span style="font-weight: 400;">Immunoprecipitation solutions for histone and transcription factor ChIP-qPCR and ChIP-seq for low inputs, plants, and animals including automated solutions</span><strong><span style="font-weight: 400;"><b><span style="font-weight: 400;">. <a href="../categories/chromatin-immunoprecipitation">Read more</a></span></b><br /></span></strong></p>
<p style="padding-left: 30px;"><strong><img src="https://www.diagenode.com/img/applications/ChIPmentation.png" alt="" width="35" height="35" /> ChIPmentation<br /><span style="font-weight: 400;">ChIPmentation, an exclusive technology, is an end-to-end ChIP-seq solution for low and difficult inputs. <a href="../categories/chromatin-ip-chipmentation">Read more</a></span><br /></strong></p>
<p style="padding-left: 30px;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/ChIP-antibodies.png" alt="" width="35" height="35" /> ChIP and ChIP-seq antibodies<br /></b></span></strong><span style="font-weight: 400;">ChIP-grade antibodies are essential for success. <a href="../categories/antibodies">Learn more</a></span><span style="font-weight: 400;"><br /></span></p>
<p style="padding-left: 30px;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/ChIP-qPCR.png" width="35" height="35" caption="false" /> Primer pairs<br /></b></span></strong><span style="font-weight: 400;">Highly specific primer pairs for the amplification of the specific genomic regions. <a href="../categories/primer-pairs">Read more</a></span><span style="font-weight: 400;"><br /></span></p>
<p style="padding-left: 30px;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/cutandtag.png" width="35" height="35" caption="false" /> CUT&Tag solutions<br /></b></span></strong><span style="font-weight: 400;">An alternative to ChIP-seq that combines antibody-targeted controlled cleavage by a protein A-Tn5 fusion with NGS to identify the binding sites of DNA-associated proteins. <a href="../categories/cutandtag">Read more</a></span><span style="font-weight: 400;"><br /></span></p>
<p style="padding-left: 30px;"><span style="font-weight: 400;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/dna-purification.png" width="35" height="35" caption="false" /> DNA purification<br /></b></span></strong><span style="font-weight: 400;"><a href="../categories/dna-and-rna-purification">Read more</a> about solutions for DNA purification</span></span></p>
<p style="padding-left: 30px;"><span style="font-weight: 400;"><span style="font-weight: 400;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/ChIP-seq.png" width="35" height="35" caption="false" /> Library preparation for ChIP-seq<br /></b></span></strong><span style="font-weight: 400;">Optimized solutions for the library preparation from low DNA input. <a href="../categories/library-preparation-for-ChIP-seq">Read more</a></span></span></span></p>
<p style="padding-left: 30px;"><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/ChIP-automation.png" alt="" width="35" height="35" /> ChIP and ChIP-seq automation<br /></b>Reproducibility, optimization simplicity, no variability. <a href="../categories/epigenetic-automation">Learn more</a></span></p>
<div class="row" style="margin-top: 32px;">
<div class="small-12 medium-10 large-9 small-centered columns">
<div class="radius panel" style="background-color: #fff;">
<p class="text-justify">We offer <a href="https://www.diagenode.com/en/categories/chromatin-immunoprecipitation" target="_blank">complete ChIP kits</a> or <strong>individual kit components</strong> from antibodies, buffers, beads, chromatin shearing, and purification reagents. With the ChIP Kit Customizer, you have complete flexibility on which components you want from our validated ChIP kits.</p>
<div class="center" style="text-align: center;"><a href="../pages/chip-kit-customizer-1"><img src="https://www.diagenode.com/img/banners/banner-customizer.png" alt="" /></a></div>
</div>
</div>
</div>
<h4>Chromatin resources</h4>
<h3>Posters</h3>
<ul>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/antibody-chipseq-qc-using-the-ipstar-compact-poster"><span style="font-weight: 400;">Understanding our antibody QC</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/antibodies-you-can-trust-poster"><span style="font-weight: 400;">High quality ChIP antibodies</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/chip-kit-results-with-true-microchip-kit-poster"><span style="font-weight: 400;">ChIP with only 10,000 cells</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/high-resolution-chipseq-profiles-with-ipstar-automated-platform-poster"><span style="font-weight: 400;">High resolution ChIP-seq using automation</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/bioinformatics-pipeline-for-chipseq-analyses"><span style="font-weight: 400;">ChIP-seq bioinformatics</span></a></li>
</ul>
<h3><span style="font-weight: 400;">Application notes</span></h3>
<ul>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/chipettor-application-note"><span style="font-weight: 400;">Simple semi-automaton for easy and inexpensive ChIP</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/chipseq-from-human-tumor-tissue"><span style="font-weight: 400;">Performing ChIP-seq on human tumor tissue</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/plant-chip-seq-application-note"><span style="font-weight: 400;">Plant ChIP-seq – a successful method</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/chip-seq-application-note"><span style="font-weight: 400;">Best workflow practices for low input ChIP</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/files/application_notes/AN-ChIP-Cas9-02_2018.pdf"><span style="font-weight: 400;">Optimize the selection of guide RNA by ChIP to keep CRISPR on-target</span></a></li>
</ul>
<h3>Publications related to ChIP</h3>
<ul>
<li><a href="https://www.diagenode.com/en/publications/view/3373">Corticosteroid receptors adopt distinct cyclical transcriptional signatures</a></li>
<li><a href="https://www.diagenode.com/en/publications/view/3347">Pro-inflammatory cytokine and high doses of ionizing radiation have similar effects on the expression of NF-kappaB-dependent genes</a></li>
<li><a href="https://www.diagenode.com/en/publications/view/3355">Functional dissection of Drosophila melanogaster SUUR protein influence on H3K27me3 profile</a></li>
</ul>
<h3><span style="font-weight: 400;">Brochures</span></h3>
<ul>
<li><a href="https://www.diagenode.com/files/brochures/Chromatin_Immunoprecipitation_Brochure.pdf"><b>Chromatin </b><span style="font-weight: 400;">products brochure</span></a></li>
<li><a href="https://www.diagenode.com/files/brochures/Epigenetic_Antibodies_Brochure.pdf"><span style="font-weight: 400;">Epigenetic<span> </span></span><b>Antibodies</b></a></li>
<li><a href="https://www.diagenode.com/files/brochures/Bioruptor_Sonicator_Brochure.pdf"><span style="font-weight: 400;">Bioruptor for<span> </span></span><b>chromatin shearing</b></a></li>
<li><a href="https://www.diagenode.com/files/brochures/IPStar_Automated_System_Brochure.pdf"><span style="font-weight: 400;">Automating<span> </span></span><b>ChIP</b><span style="font-weight: 400;"><span> </span>and<span> </span></span><b>ChIP-seq</b></a></li>
</ul>
</div>
</div>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => true,
'hide' => false,
'all_format' => false,
'is_antibody' => false,
'slug' => 'chromatin-function',
'cookies_tag_id' => null,
'meta_keywords' => 'Chromatin function,Chromatin shearing,Chromatin immunoprecipitation,Chromatin assembly',
'meta_description' => 'Diagenode offers a number of unique solutions and kits to make Diagenode offers chromatin immunoprecipitation kits have been designed to meet a wide variety of research needs and accessible',
'meta_title' => 'Chromatin Function Kit for Unique Solutions and Kits to make Chromatin Research| Diagenode',
'modified' => '2025-05-21 12:06:52',
'created' => '2015-05-27 05:22:15',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
)
),
'Document' => array(
(int) 0 => array(
'id' => '1139',
'name' => 'ATAC-seq Kit - Manual',
'description' => '<p class="p1">Gene expression is carefully regulated in the cells in order to manage a wide range of biological functions. The structure of chromatin is quite dynamic and contributes to this crucial regulatory process.</p>
<p class="p1">ATAC-seq, Assay for Transposase-Accessible Chromatin, followed by nextgeneration sequencing, is a key technology to easily identify the “open” regions of the chromatin, which are usually associated with permissive gene expression. Indeed, the nuclei of the samples are incubated with a transposase, and only the genomic regions associated with open chromatin will be accessible to this transposase. During the process those regions will be cut and sequencing adaptors will be added, allowing their sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only., giving a map of the chromatin status in the whole genome of the sample.</p>
<p class="p1">The Diagenode’s ATAC-seq kit is based on a highly validated protocol, used for years in our Epigenomics Profiling Services offer and takes advantage of many successful Diagenode’s tools, such as the loaded Tagmentase (Tn5 transposase), the MicroChIP DiaPure Columns and the Primer indexes for tagmented libraries kits.</p>',
'image_id' => null,
'type' => 'Manual',
'url' => 'files/products/kits/atacseq-kit-manual.pdf',
'slug' => 'atacseq-kit-manual',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-06-25 09:51:59',
'created' => '2021-06-25 09:51:59',
'ProductsDocument' => array(
[maximum depth reached]
)
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
'id' => '1819',
'name' => 'product/kits/atacseq-kit-picture.jpg',
'alt' => 'ATAC-seq kit',
'modified' => '2021-06-25 10:03:22',
'created' => '2021-06-25 10:03:22',
'ProductsImage' => array(
[maximum depth reached]
)
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
'id' => '5143',
'name' => 'CRAMP1 drives linker histone expression to enable Polycomb repression',
'authors' => 'Matthews, Rachael E et al.',
'description' => '<div class="abstract-content selected" id="eng-abstract">
<p style="text-align: justify;">In contrast to the well-understood role of core histones in DNA packaging, the function of the linker histone (H1) remains enigmatic. Challenging the prevailing view that linker histones are a general feature of heterochromatin, here we show a critical requirement for H1 in Polycomb repressive complex 2 (PRC2) function. A CRISPR-Cas9 genetic screen using a fluorescent PRC2 reporter identified an essential role for the poorly characterized gene CRAMP1 in PRC2-mediated repression. CRAMP1 localizes to the promoters of expressed H1 genes and positively regulates their transcription. CRAMP1 ablation simultaneously depletes all linker histones, which results in selective decompaction of H3K27me3-marked loci and derepression of PRC2 target genes without concomitant loss of PRC2 occupancy or enzymatic activity. Strikingly, we find that linker histones preferentially localize to genomic loci marked by H3K27me3 across diverse cell types and organisms. Altogether, these data demonstrate a prominent role for linker histones in epigenetic repression by PRC2.</p>
</div>',
'date' => '2025-06-10',
'pmid' => 'https://pubmed.ncbi.nlm.nih.gov/40516528/',
'doi' => '10.1016/j.molcel.2025.05.031',
'modified' => '2025-06-18 09:45:01',
'created' => '2025-06-18 09:45:01',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '5142',
'name' => 'Temporal and spatial niche partitioning in a retrotransposon community of the Drosophila melanogaster genome',
'authors' => 'Varoqui, Marion et al.',
'description' => '<div class="abstract" id="abstract">
<div class="abstract-content selected" id="eng-abstract">
<p style="text-align: justify;">Transposable elements (TEs) are genetic parasites that can potentially threaten the stability of the genomes they colonize. Nonetheless, TEs persist within genomes and are rarely fully eliminated, with diverse TE families coexisting in varing copy numbers. The TE replication strategies that enable host organisms to tolerate and accommodate the extensive diversity of TEs, while minimizing harm to the host and avoiding mutual competition among TEs, remain poorly understood. Here, by studying the spontaneous or experimental mobilization of four Drosophila LTR RetroTransposable Elements (LTR-RTEs), we reveal that each of them preferentially targets open chromatin regions characterized by specific epigenetic features. Among these, gtwin and ZAM are expressed in distinct cell types within female somatic gonadal tissues and inserted into the distinct accessible chromatin landscapes of the corresponding stages of embryogenesis. These findings suggest that individual LTR-RTEs exploit unique biological niches, enabling their coexistence within the tightly regulated ecosystem of the same host genome.</p>
</div>
</div>',
'date' => '2025-06-06',
'pmid' => 'https://pubmed.ncbi.nlm.nih.gov/40503684/',
'doi' => '10.1093/nar/gkaf516',
'modified' => '2025-06-18 09:43:08',
'created' => '2025-06-18 09:43:08',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '5128',
'name' => 'Cervicovaginal microbiome alters transcriptomic and epigenomic signatures across cervicovaginal epithelial barriers',
'authors' => 'Lauren Anton et al. ',
'description' => '<p><b>Background</b></p>
<p>The cervicovaginal microbiome plays a critical role in women's health, with microbial communities dominated by<span> </span><em>Lactobacillus</em><span> </span>species considered optimal. In contrast, the depletion of lactobacilli and the presence of a diverse array of strict and facultative anaerobes, such as<span> </span><em>Gardnerella vaginalis</em>, have been linked with adverse reproductive outcomes. Despite these associations, the molecular mechanisms by which host-microbial interactions modulate cervical and vaginal epithelial function remains poorly understood.</p>
<p><b>Results</b></p>
<p>In this study, we used RNA sequencing to characterize the transcriptional response of cervicovaginal epithelial cells exposed to the culture supernatants of common vaginal bacteria. Our findings revealed that<span> </span><em>G. vaginalis</em><span> </span>culture supernatants upregulate genes associated with an activated innate immune response and increased cell death. Conversely,<span> </span><em>Lactobacillus crispatus</em><span> </span>culture supernatants induced transcriptional changes indicative of epigenomic modeling in ectocervical epithelial cells. Epigenomic modification by<span> </span><em>L. crispatus</em>, was confirmed by ATAC-sequencing, which demonstrated reduced chromatin accessibility.</p>
<p><b>Conclusions</b></p>
<p>These results provide new insights into host-microbe interactions within the lower reproductive tract and suggests that modulating the vaginal microbiome could offer innovative therapeutic strategies to improve reproductive health.</p>',
'date' => '2025-05-07',
'pmid' => 'https://www.researchsquare.com/article/rs-6171614/v1',
'doi' => 'https://doi.org/10.21203/rs.3.rs-6171614/v1',
'modified' => '2025-05-12 10:30:24',
'created' => '2025-05-12 10:30:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '5125',
'name' => 'An ISWI-related chromatin remodeller regulates stage-specific gene expression in Toxoplasma gondii',
'authors' => 'Pachano, Belen et al.',
'description' => '<p style="text-align: justify;"><span>ATP-dependent chromatin remodellers are specialized multiprotein machines that organize the genome in eukaryotic cells and regulate its accessibility by repositioning, ejecting or modifying nucleosomes. However, their role in </span><i>Toxoplasma gondii</i><span><span> </span>is poorly understood. Here we show that<span> </span></span><i>T.</i><span> </span><i>gondii</i><span><span> </span>has evolved two divergent proteins within the imitation switch (ISWI) family:<span> </span></span><i>Tg</i><span>SNF2h and<span> </span></span><i>Tg</i><span>SNF2L.<span> </span></span><i>Tg</i><span>SNF2h specifically forms a core complex with the transcription factor AP2VIII-2 and the scaffold protein<span> </span></span><i>Tg</i><span>RFTS. Depletion of<span> </span></span><i>Tg</i><span>RFTS phenocopies the knockdown of<span> </span></span><i>Tg</i><span>SNF2h, restricting access to chromatin and altering local gene expression. At the genomic level,<span> </span></span><i>Tg</i><span>SNF2h insulates highly transcribed genes from silenced neighbours, ensuring stage-specific gene regulation. By modulating chromatin accessibility to transcription factors,<span> </span></span><i>Tg</i><span>SNF2h exerts epistatic control over MORC, a key regulator of sexual commitment. Our findings show that a specific ISWI complex orchestrates the partitioning of developmental genes and ensures transcriptional fidelity throughout the parasite life cycle.</span></p>',
'date' => '2025-04-11',
'pmid' => 'https://www.nature.com/articles/s41564-025-01980-2#citeas',
'doi' => 'https://doi.org/10.1038/s41564-025-01980-2',
'modified' => '2025-05-19 11:36:47',
'created' => '2025-05-06 15:49:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '5101',
'name' => 'Repression of oxidative phosphorylation by NR2F2, MTERF3 and GDF15 in human skin under high-glucose stress',
'authors' => 'S. Ley-Ngardigal et al.',
'description' => '<p><span>Lifestyle factors such as a Western diet or metabolic diseases like diabetes disrupt glucose homeostasis and induce stress responses, yet their impact on skin metabolism and structural integrity remains poorly understood. Here, we performed multiomic and bioenergetic analyses of human dermal fibroblasts (HDFs), human equivalent dermis (HED), human reconstructed skin (HRS), and skin explants from diabetic patients. We found that 12 mM glucose stress represses oxidative phosphorylation (OXPHOS) through a dual mechanism: the glucose-dependent nuclear receptor NR2F2 activates mitochondrial transcription termination factor 3 (MTERF3) while inhibiting growth-differentiation factor 15 (GDF15). Promoter assays revealed that MTERF3 is regulated by NR2F2 and MYCN, whereas GDF15 is modulated by NR2F2 and FOS. Consequently, OXPHOS proteins and mitochondrial respiration were suppressed, and MTERF3 overexpression additionally interfered with collagen biosynthesis. In contrast, GDF15 supplementation fully rescued hyperglycemia-induced bioenergetic and metabolomic alterations, suggesting a pharmacological strategy to mitigate hyperglycemic damage in the skin. Finally, silencing GDF15 or TFAM impaired fibroblast haptotaxis and skin reconstruction, underscoring the crucial role of mitochondrial energetics in dermal structure and function. Collectively, these findings identify the NR2F2–MTERF3–GDF15 axis as a key mediator of OXPHOS suppression and highlight a potential therapeutic target to preserve skin integrity under hyperglycemic stress.</span></p>',
'date' => '2025-03-27',
'pmid' => 'https://www.sciencedirect.com/science/article/pii/S2213231725001260',
'doi' => 'https://doi.org/10.1016/j.redox.2025.103613',
'modified' => '2025-03-31 13:44:10',
'created' => '2025-03-31 13:44:10',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '5092',
'name' => 'CRISPR screening identifies regulators of enhancer-mediated androgen receptor transcription in advanced prostate cancer',
'authors' => 'Rachel R. Xiang et al. ',
'description' => '<section id="author-highlights-abstract" property="abstract" typeof="Text" role="doc-abstract">
<h2 property="name"></h2>
</section>
<section id="author-abstract" property="abstract" typeof="Text" role="doc-abstract">
<div id="abspara0010" role="paragraph">Amplification of the androgen receptor (AR) locus is the most frequent alteration in metastatic castration-resistant prostate cancer (CRPC). Recently, it was discovered that an enhancer of the AR is co-amplified with the AR gene body and contributes to increased AR transcription and resistance to androgen deprivation therapy. However, the mechanism of enhancer activation in advanced disease is unknown. Here, we used CRISPR-Cas9 screening to identify transcription factors that bind to the AR enhancer and modulate enhancer-mediated AR transcription. We demonstrate that HOXB13, GATA2, and TFAP2C bind the AR enhancer in patient-derived xenografts and directly impact features associated with an active chromatin state. Interestingly, the AR enhancer belongs to a set of regulatory elements that require HOXB13 to maintain FOXA1 binding, further delineating the role of HOXB13 in CRPC. This work provides a framework to functionally identify<span> </span><i>trans</i>-acting factors required for the activation of disease-related noncoding regulatory elements.</div>
</section>',
'date' => '2025-02-14',
'pmid' => 'https://www.cell.com/cell-reports/fulltext/S2211-1247(25)00083-X',
'doi' => '10.1016/j.celrep.2025.115312',
'modified' => '2025-03-27 16:56:52',
'created' => '2025-03-27 16:54:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '5038',
'name' => 'Schistosoma mansoni antigen induced innate immune memory features mitochondrial biogenesis and can be inhibited by ovarian produced hormones',
'authors' => 'Juan Marcos Oviedo et al.',
'description' => '<p><span>We have previously identified that </span><em>S. mansoni</em><span><span> </span>infection induces a unique form of myeloid training that protects male but not female mice from high fat diet induced disease. Here we demonstrate that ovarian derived hormones account for this sex specific difference. Ovariectomy of females prior to infection permits metabolic reprogramming of the myeloid lineage, with BMDM exhibiting carbon source flexibility for cellular respiration, and mice protected from systemic metabolic disease. The innate training phenotype of infection can be replicated by<span> </span></span><em>in vivo</em><span><span> </span>injection of SEA, and by exposure of bone marrow to SEA in culture prior to macrophage differentiation (Day 0). This protective phenotype is linked to increased chromatin accessibility of lipid and mitochondrial pathways in BMDM including Nrf1 and Tfam, as well as mitochondrial biogenesis. This work provides evidence that<span> </span></span><em>S. mansoni</em><span><span> </span>antigens induce a unique form of innate training inhibited by ovarian-derived hormones in females.</span></p>',
'date' => '2025-01-18',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2025.01.14.632838v1',
'doi' => 'https://doi.org/10.1101/2025.01.14.632838',
'modified' => '2025-01-31 10:55:42',
'created' => '2025-01-31 10:55:42',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '5117',
'name' => 'Dynamic PRC1–CBX8 stabilizes a porous structure of chromatin condensates',
'authors' => 'Uckelmann, M., et al.',
'description' => '<p><strong>Abstract</strong></p>
<div class="c-article-section__content" id="Abs1-content">
<p style="text-align: justify;">The compaction of chromatin is a prevalent paradigm in gene repression. Chromatin compaction is commonly thought to repress transcription by restricting chromatin accessibility. However, the spatial organization and dynamics of chromatin compacted by gene-repressing factors are unknown. Here, using cryo-electron tomography, we solved the three-dimensional structure of chromatin condensed by the polycomb repressive complex 1 (PRC1) in a complex with CBX8. PRC1-condensed chromatin is porous and stabilized through multivalent dynamic interactions of PRC1 with chromatin. Mechanistically, positively charged residues on the internally disordered regions of CBX8 mask negative charges on the DNA to stabilize the condensed state of chromatin. Within condensates, PRC1 remains dynamic while maintaining a static chromatin structure. In differentiated mouse embryonic stem cells, CBX8-bound chromatin remains accessible. These findings challenge the idea of rigidly compacted polycomb domains and instead provide a mechanistic framework for dynamic and accessible PRC1–chromatin condensates.</p>
</div>',
'date' => '2025-01-15',
'pmid' => 'https://www.nature.com/articles/s41594-024-01457-6',
'doi' => 'https://doi.org/10.1038/s41594-024-01457-6',
'modified' => '2025-04-25 13:20:40',
'created' => '2025-04-25 11:48:09',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 8 => array(
'id' => '5076',
'name' => 'Assessing the impact of whole genome duplication on gene expression and regulation during arachnid development',
'authors' => 'Madeleine E. Aase-Remedios et al.',
'description' => '<p id="p-2">Whole genome duplication (WGD) generates new genetic material that can contribute to the evolution of the regulation of developmental processes and phenotypic diversification. A WGD occurred in an ancestor of arachnopulmonates (spiders, scorpions, and their relatives), which provides an important independent comparison to WGDs in other animal lineages like those in vertebrates and horseshoe crabs. After WGD, arachnopulmonates retained many duplicated copies (ohnologues) of developmental genes including clusters of homeobox genes, many of which have been inferred to have undergone subfunctionalisation. However, there has been little systematic analysis of gene regulatory sequences and comparison of the expression of ohnologues versus their single-copy orthologues between arachnids. Here we compare the regions of accessible chromatin and gene expression of ohnologues and single-copy genes during three embryonic stages between an arachnopulmonate arachnid, the spider<span> </span><em>Parasteatoda tepidariorum</em>, and a non-arachnopulmonate arachnid, the harvestman<span> </span><em>Phalangium opilio</em>. We found that the expression of each spider ohnologue was lower than their single-copy orthologues in the harvestman providing evidence for subfunctionalisation. However, this was not reflected in a reduction in peaks of accessible chromatin because both spider ohnologues and single-copy genes had more peaks than the harvestman genes. We also found peaks of open chromatin that increased in the late stage associated with activation of genes expressed later during embryogenesis in both species. Taken together, our study provides the first genome-wide comparison of gene regulatory sequences and gene expression in arachnids and thus provides new insights into the impact of the arachnopulmonate WGD.</p>
<div id="sec-1" class="subsection">
<p id="p-3"><strong>Significance statement</strong><span> </span>The comparison of independent WGD events is essential to understanding their evolutionary outcomes. Here we examine gene expression and chromatin accessibility during the development of two arachnids, one of which underwent an ancient WGD. Our data provide the first direct comparison of the regulation of ohnologues and single-copy genes in arachnids.</p>
</div>',
'date' => '2024-12-20',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2024.12.18.628675v1.full',
'doi' => 'https://doi.org/10.1101/2024.12.18.628675',
'modified' => '2025-02-27 17:24:20',
'created' => '2025-02-27 17:24:20',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 9 => array(
'id' => '5012',
'name' => 'Spatial organizations of heterochromatin underpin nuclear structural integrity of ventricular cardiomyocytes against mechanical stress',
'authors' => 'Keita Fujiwara et al.',
'description' => '<section id="author-highlights-abstract" property="abstract" typeof="Text" role="doc-abstract">
<h2 property="name">Highlights</h2>
<div id="abspara0020" role="paragraph">
<div id="ulist0010" role="list">
<div id="u0010" role="listitem">
<div class="content">
<div id="p0010" role="paragraph">Cardiomyocytes acquire characteristic spatial organizations of heterochromatin (SOH)</div>
</div>
</div>
<div id="u0015" role="listitem">
<div class="content">
<div id="p0015" role="paragraph">High levels of H2B-mCherry disrupt SOH, leading to nuclear elongation in cardiomyocytes</div>
</div>
</div>
<div id="u0020" role="listitem">
<div class="content">
<div id="p0020" role="paragraph">SOH disruption minimally impacts gene expression despite loosening global genome structure</div>
</div>
</div>
<div id="u0025" role="listitem">
<div class="content">
<div id="p0025" role="paragraph">SOH alter with aging, leading to nuclear deformation in ventricular cardiomyocytes</div>
</div>
</div>
</div>
</div>
</section>
<section id="author-abstract" property="abstract" typeof="Text" role="doc-abstract">
<h2 property="name">Summary</h2>
<div id="abspara0010" role="paragraph">Cardiomyocyte (CM) nuclei are constantly exposed to mechanical stress, but how they maintain their nuclear shape remains unknown. In this study, we found that ventricular CM nuclei acquire characteristic prominent spatial organizations of heterochromatin (SOH), which are disrupted by high-level expression of H2B-mCherry in mice. SOH disruption was associated with nuclear softening, leading to extreme elongation and rupture under unidirectional mechanical stress. Loosened chromatin then leaks into the cytosol, causing severe inflammation and cardiac dysfunction. Although SOH disruption was accompanied by loosened higher-order genomic structures, the change in gene expression before nuclear deformation was mild, suggesting that SOH play major roles in nuclear structural integrity. Aged CM nuclei consistently exhibited scattered SOH and marked elongation. Furthermore, we provide mechanistic insight into the development and maintenance of SOH driven by chromatin compaction and condensate formation. These results highlight SOH as a safeguard of nuclear shape and genomic integrity against mechanical stress.</div>
</section>',
'date' => '2024-12-09',
'pmid' => 'https://www.cell.com/cell-reports/fulltext/S2211-1247(24)01399-8',
'doi' => '10.1016/j.celrep.2024.115048',
'modified' => '2024-12-12 15:04:47',
'created' => '2024-12-12 15:04:47',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 10 => array(
'id' => '4984',
'name' => 'DNA demethylation triggers cell free DNA release in colorectal cancer cells',
'authors' => 'Valeria Pessei et al.',
'description' => '<h3 class="c-article__sub-heading" data-test="abstract-sub-heading">Background</h3>
<p>Liquid biopsy based on cell-free DNA (cfDNA) analysis holds significant promise as a minimally invasive approach for the diagnosis, genotyping, and monitoring of solid malignancies. Human tumors release cfDNA in the bloodstream through a combination of events, including cell death, active and passive release. However, the precise mechanisms leading to cfDNA shedding remain to be characterized. Addressing this question in patients is confounded by several factors, such as tumor burden extent, anatomical and vasculature barriers, and release of nucleic acids from normal cells. In this work, we exploited cancer models to dissect basic mechanisms of DNA release.</p>
<h3 class="c-article__sub-heading" data-test="abstract-sub-heading">Methods</h3>
<p>We measured cell loss ratio, doubling time, and cfDNA release in the supernatant of a colorectal cancer (CRC) cell line collection (<i>N</i> = 76) representative of the molecular subtypes previously identified in cancer patients. Association analyses between quantitative parameters of cfDNA release, cell proliferation, and molecular features were evaluated. Functional experiments were performed to test the impact of modulating DNA methylation on cfDNA release.</p>
<h3 class="c-article__sub-heading" data-test="abstract-sub-heading">Results</h3>
<p>Higher levels of supernatant cfDNA were significantly associated with slower cell cycling and increased cell death. In addition, a higher cfDNA shedding was found in non-CpG Island Methylator Phenotype (CIMP) models. These results indicate a positive correlation between lower methylation and increased cfDNA levels. To explore this further, we exploited methylation microarrays to identify a subset of probes significantly associated with cfDNA shedding and derive a methylation signature capable of discriminating high from low cfDNA releasers. We applied this signature to an independent set of 176 CRC cell lines and patient derived organoids to select 14 models predicted to be low or high releasers. The methylation profile successfully predicted the amount of cfDNA released in the supernatant. At the functional level, genetic ablation of DNA methyl-transferases increased chromatin accessibility and DNA fragmentation, leading to increased cfDNA release in isogenic CRC cell lines. Furthermore, in vitro treatment of five low releaser CRC cells with a demethylating agent was able to induce a significant increase in cfDNA shedding.</p>
<h3 class="c-article__sub-heading" data-test="abstract-sub-heading">Conclusions</h3>
<p>Methylation status of cancer cell lines contributes to the variability of cfDNA shedding in vitro. Changes in methylation pattern are associated with cfDNA release levels and might be exploited to increase sensitivity of liquid biopsy assays.</p>',
'date' => '2024-10-09',
'pmid' => 'https://link.springer.com/article/10.1186/s13073-024-01386-5',
'doi' => 'https://doi.org/10.1186/s13073-024-01386-5',
'modified' => '2024-10-14 08:56:24',
'created' => '2024-10-14 08:56:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 11 => array(
'id' => '4967',
'name' => 'Temporal and spatial niche partitioning in a retrotransposon community of the Drosophila genome',
'authors' => 'Varoqui M. et al.',
'description' => '<p><span>Transposable elements (TEs), widespread genetic parasites, pose potential threats to the stability of their host genomes. Hence, the interactions observed today between TEs and their host genomes, as well as among the different TE species coexisting in the same host, likely reflect those that did not lead to the extinction of either the host or the TEs. It is not clear to what extent the expression and integration steps of the TE replication cycles are involved in this ‘peaceful’ coexistence. Here, we show that four Drosophila LTR RetroTransposable Elements (LTR-RTEs), although sharing the same overall integration mechanism, preferentially integrate into distinct open chromatin domains of the host germline. Notably, the differential expressions of the gtwin and ZAM LTR-RTEs in ovarian and embryonic somatic tissues, respectively, result in differential integration timings and targeting of accessible chromatin landscapes that differ between early and late embryonic nuclei, highlighting connections between temporal and spatial LTR-RTEs niche partitionings.</span></p>',
'date' => '2024-08-16',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2024.08.14.607943v1',
'doi' => 'https://doi.org/10.1101/2024.08.14.607943',
'modified' => '2024-09-02 10:29:44',
'created' => '2024-09-02 10:29:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 12 => array(
'id' => '5093',
'name' => 'Galectin 3-binding protein (LGALS3BP) depletion attenuates hepatic fibrosis by reducing transforming growth factor-β1 (TGF-β1) availability and inhibits hepatocarcinogenesis',
'authors' => 'Dae-Hwan Kim et al.',
'description' => '<section class="article-section__content" id="cac212600-sec-0010">
<h3 class="article-section__sub-title section1" id="cac212600-sec-0010-title">Background</h3>
<p>Increased Galectin 3-binding protein (LGALS3BP) serum levels have been used to assess hepatic fibrosis stages and the severity of hepatocellular carcinoma (HCC). Considering the crucial role of transforming growth factor-β1 (TGF-β1) in the emergence of these diseases, the present study tested the hypothesis that LGALS3BP regulates the TGF-β1 signaling pathway.</p>
</section>
<section class="article-section__content" id="cac212600-sec-0020">
<h3 class="article-section__sub-title section1" id="cac212600-sec-0020-title">Methods</h3>
<p>The expression levels of<span> </span><i>LGALS3BP</i><span> </span>and<span> </span><i>TGFB1</i><span> </span>were analyzed in patients with metabolic dysfunction-associated steatohepatitis (MASH) and HCC. Multiple omics techniques, such as RNA-sequencing, transposase-accessible chromatin-sequencing assay, and liquid chromatography-tandem mass spectrometry proteomics, were used to identify the regulatory mechanisms for the LGALS3BP-TGF-β1 axis. The effects of altered TGF-β1 signaling by LGALS3BP were investigated in conditional<span> </span><i>LGALS3BP</i>-knockin and<span> </span><i>LGALS3BP</i>-knockout mice.</p>
</section>
<section class="article-section__content" id="cac212600-sec-0030">
<h3 class="article-section__sub-title section1" id="cac212600-sec-0030-title">Results</h3>
<p>In patients with MASH and HCC, the levels of<span> </span><i>LGALS3BP</i><span> </span>and<span> </span><i>TGFB1</i><span> </span>exhibited positive correlations. Stimulation of LGALS3BP by the inflammatory cytokine interferon α in HCC cells or ectopic overexpression of LGALS3BP in hepatocytes promoted the expression levels of TGFB1. Aggravated fibrosis was observed in the livers of hepatocyte-specific<span> </span><i>LGALS3BP</i>-knockin mice, with increased TGFB1 levels. LGALS3BP directly bound to and assembled integrin αV, an integral mediator required for releasing active TGF-β1 from extracellular latent complex with the rearranged F-actin cytoskeleton. The released TGF-β1 activated JunB transcription factor, which in turn promoted the TGF-β1 positive feedback loop.<span> </span><i>LGALS3BP</i><span> </span>deletion in the hepatocytes downregulated TGF-β1 signaling and CCl<sub>4</sub><span> </span>induced fibrosis. Moreover,<span> </span><i>LGALS3BP</i><span> </span>depletion hindered hepatocarcinogenesis by limiting the availability of fibrogenic TGF-β1.</p>
</section>
<section class="article-section__content" id="cac212600-sec-0040">
<h3 class="article-section__sub-title section1" id="cac212600-sec-0040-title">Conclusion</h3>
<p>LGALS3BP plays a crucial role in hepatic fibrosis and carcinogenesis by controlling the TGF-β1 signaling pathway, making it a promising therapeutic target in TGF-β1-related diseases.</p>
</section>',
'date' => '2024-07-28',
'pmid' => 'https://onlinelibrary.wiley.com/doi/full/10.1002/cac2.12600',
'doi' => ' https://doi.org/10.1002/cac2.12600',
'modified' => '2025-03-27 16:59:26',
'created' => '2025-03-27 16:59:26',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 13 => array(
'id' => '5091',
'name' => 'Patho-transcriptomic analysis of invasive mucinous adenocarcinoma of the lung (IMA): comparison with lung adenocarcinoma with signet ring cell features (SRCC)',
'authors' => 'William D. Stuart et al.',
'description' => '<div id="sec-1" class="subsection">
<p id="p-3"><strong>Background</strong><span> </span>Invasive mucinous adenocarcinoma (IMA) comprises ∼5% of lung adenocarcinoma. There is no effective therapy for IMA when surgical resection is not possible. IMA is sometimes confused with adenocarcinoma with signet ring cell features (SRCC) pathologically since both adenocarcinomas feature tumor cells with abundant intracellular mucin. The molecular mechanisms by which such mucin-producing lung adenocarcinomas develop remain unknown.</p>
</div>
<div id="sec-2" class="subsection">
<p id="p-4"><strong>Methods</strong><span> </span>Using a Visium spatial transcriptomics approach, we analyzed IMA and compared it with SRCC patho-transcriptomically. Combining spatial transcriptomics data with<span> </span><em>in vitro</em><span> </span>studies using RNA-seq and ChIP-seq, we assessed downstream targets of transcription factors HNF4A and SPDEF that are highly expressed in IMA and/or SRCC.</p>
</div>
<div id="sec-3" class="subsection">
<p id="p-5"><strong>Results</strong><span> </span>Spatial transcriptomics analysis indicated that there are 6 distinct cell clusters in IMA and SRCC. Notably, two clusters (C1 and C3) of mucinous tumor cells exist in both adenocarcinomas albeit at a different ratio. Importantly, a portion of genes (e.g.,<span> </span><em>NKX2-1</em>,<span> </span><em>GKN1</em>,<span> </span><em>HNF4A</em><span> </span>and<span> </span><em>FOXA3</em>) are distinctly expressed while some mucous-related genes (e.g.,<span> </span><em>SPDEF</em><span> </span>and<span> </span><em>FOXA2</em>) are expressed in both adenocarcinomas. We determined that HNF4A induces<span> </span><em>MUC3A/B</em><span> </span>and<span> </span><em>TM4SF4</em><span> </span>and that BI 6015, an HNF4A antagonist, suppressed the growth of IMA cells. Using mutant SPDEF that is associated with COVID-19, we also determined that an intact DNA-binding domain of SPDEF is required for SPDEF-mediated induction of mucin genes (<em>MUC5AC</em>,<span> </span><em>MUC5B</em><span> </span>and<span> </span><em>AGR2</em>). Additionally, we found that XMU-MP-1, a SPDEF inhibitor, suppressed the growth of IMA cells.</p>
</div>
<div id="sec-4" class="subsection">
<p id="p-6"><strong>Conclusion</strong><span> </span>These results revealed that IMA and SRCC contain heterogenous tumor cell types, some of which are targetable.</p>
</div>',
'date' => '2024-06-17',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2024.06.13.598839v1.full',
'doi' => 'https://doi.org/10.1101/2024.06.13.598839',
'modified' => '2025-03-27 16:08:19',
'created' => '2025-03-27 16:08:19',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 14 => array(
'id' => '5094',
'name' => 'Transient loss of Polycomb components induces an epigenetic cancer fate',
'authors' => 'V. Parreno et al.',
'description' => '<p><span>Although cancer initiation and progression are generally associated with the accumulation of somatic mutations</span><sup><a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 1" title="McGranahan, N. & Swanton, C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 27, 15–26 (2015)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR1" id="ref-link-section-d121858e719">1</a>,<a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 2" title="Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR2" id="ref-link-section-d121858e722">2</a></sup><span>, substantial epigenomic alterations underlie many aspects of tumorigenesis and cancer susceptibility</span><sup><a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" title="Brock, A., Chang, H. & Huang, S. Non-genetic heterogeneity—a mutation-independent driving force for the somatic evolution of tumours. Nat. Rev. Genet. 10, 336–342 (2009)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR3" id="ref-link-section-d121858e726">3</a>,<a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" title="Flavahan, W. A., Gaskell, E. & Bernstein, B. E. Epigenetic plasticity and the hallmarks of cancer. Science 357, eaal2380 (2017)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR4" id="ref-link-section-d121858e726_1">4</a>,<a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" title="Marine, J. C., Dawson, S. J. & Dawson, M. A. Non-genetic mechanisms of therapeutic resistance in cancer. Nat. Rev. Cancer 20, 743–756 (2020)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR5" id="ref-link-section-d121858e726_2">5</a>,<a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 6" title="Timp, W. & Feinberg, A. P. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat. Rev. Cancer 13, 497–510 (2013)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR6" id="ref-link-section-d121858e729">6</a></sup><span>, suggesting that genetic mechanisms might not be the only drivers of malignant transformation</span><sup><a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 7" title="Teixeira, V. H. et al. Deciphering the genomic, epigenomic, and transcriptomic landscapes of pre-invasive lung cancer lesions. Nat. Med. 25, 517–525 (2019)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR7" id="ref-link-section-d121858e733">7</a></sup><span>. However, whether purely non-genetic mechanisms are sufficient to initiate tumorigenesis irrespective of mutations has been unknown. Here, we show that a transient perturbation of transcriptional silencing mediated by Polycomb group proteins is sufficient to induce an irreversible switch to a cancer cell fate in<span> </span></span><i>Drosophila</i><span>. This is linked to the irreversible derepression of genes that can drive tumorigenesis, including members of the JAK–STAT signalling pathway and<span> </span></span><i>zfh1</i><span>, the fly homologue of the<span> </span></span><i>ZEB1</i><span><span> </span>oncogene, whose aberrant activation is required for Polycomb perturbation-induced tumorigenesis. These data show that a reversible depletion of Polycomb proteins can induce cancer in the absence of driver mutations, suggesting that tumours can emerge through epigenetic dysregulation leading to inheritance of altered cell fates.</span></p>',
'date' => '2024-04-24',
'pmid' => 'https://www.nature.com/articles/s41586-024-07328-w',
'doi' => 'https://doi.org/10.1038/s41586-024-07328-w',
'modified' => '2025-03-27 17:00:44',
'created' => '2025-03-27 17:00:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 15 => array(
'id' => '4933',
'name' => 'Systematic mapping of TF-mediated cell fate changes by a pooled induction coupled with scRNA-seq and multi-omics approaches',
'authors' => 'Lee M. et al.',
'description' => '<p><span>Transcriptional regulation controls cellular functions through interactions between transcription factors (TFs) and their chromosomal targets. However, understanding the fate conversion potential of multiple TFs in an inducible manner remains limited. Here, we introduce iTF-seq as a method for identifying individual TFs that can alter cell fate toward specific lineages at a single-cell level. iTF-seq enables time course monitoring of transcriptome changes, and with biotinylated individual TFs, it provides a multi-omics approach to understanding the mechanisms behind TF-mediated cell fate changes. Our iTF-seq study in mouse embryonic stem cells identified multiple TFs that trigger rapid transcriptome changes indicative of differentiation within a day of induction. Moreover, cells expressing these potent TFs often show a slower cell cycle and increased cell death. Further analysis using bioChIP-seq revealed that GCM1 and OTX2 act as pioneer factors and activators by increasing gene accessibility and activating the expression of lineage specification genes during cell fate conversion. iTF-seq has utility in both mapping cell fate conversion and understanding cell fate conversion mechanisms.</span></p>',
'date' => '2024-04-05',
'pmid' => 'https://pubmed.ncbi.nlm.nih.gov/38580401/',
'doi' => '10.1101/gr.277926.123',
'modified' => '2024-04-09 00:19:18',
'created' => '2024-04-09 00:19:18',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 16 => array(
'id' => '4895',
'name' => 'Protocol to isolate nuclei from Chlamydomonas reinhardtii for ATAC sequencing',
'authors' => 'Santhanagopalan I. et al.',
'description' => '<p class="section-title u-h4 u-margin-l-top u-margin-xs-bottom">Highlights</p>
<div id="abssec0020">
<ul class="list">
<li class="react-xocs-list-item">
<p id="p0010">Optimized isolation of nuclei from the green model alga<span> </span><em>Chlamydomonas reinhardtii</em></p>
</li>
<li class="react-xocs-list-item">
<p id="p0010"><em></em></p>
<p id="p0015">Tag-free isolation from both cell-walled and cell wall-deficient algae strains</p>
</li>
<li class="react-xocs-list-item">
<p id="p0015"></p>
<p id="p0020">Key steps for an effective and fast isolation and quantification procedure of nuclei</p>
</li>
<li class="react-xocs-list-item">
<p><span class="list-label"></span>Extracts at a quality suitable for ATAC-sequencing</p>
</li>
</ul>
</div>',
'date' => '2024-03-15',
'pmid' => 'https://www.sciencedirect.com/science/article/pii/S2666166723007311',
'doi' => 'https://doi.org/10.1016/j.xpro.2023.102764',
'modified' => '2024-02-09 12:37:32',
'created' => '2024-01-22 13:39:58',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 17 => array(
'id' => '4917',
'name' => 'Interplay between coding and non-coding regulation drives the Arabidopsis seed-to-seedling transition',
'authors' => 'Trembley B.J.M. et al.',
'description' => '<p><span>Translation of seed stored mRNAs is essential to trigger germination. However, when RNAPII re-engages RNA synthesis during the seed-to-seedling transition has remained in question. Combining csRNA-seq, ATAC-seq and smFISH in </span><i>Arabidopsis thaliana</i><span><span> </span>we demonstrate that active transcription initiation is detectable during the entire germination process. Features of non-coding regulation such as dynamic changes in chromatin accessible regions, antisense transcription, as well as bidirectional non-coding promoters are widespread throughout the Arabidopsis genome. We show that sensitivity to exogenous ABSCISIC ACID (ABA) during germination depends on proximal promoter accessibility at ABA-responsive genes. Moreover, we provide genetic validation of the existence of divergent transcription in plants. Our results reveal that active enhancer elements are transcribed producing non-coding enhancer RNAs (eRNAs) as widely documented in metazoans. In sum, this study defining the extent and role of coding and non-coding transcription during key stages of germination expands our understanding of transcriptional mechanisms underlying plant developmental transitions.</span></p>',
'date' => '2024-02-26',
'pmid' => 'https://www.nature.com/articles/s41467-024-46082-5',
'doi' => 'https://doi.org/10.1038/s41467-024-46082-5',
'modified' => '2024-02-29 11:56:56',
'created' => '2024-02-29 11:56:56',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 18 => array(
'id' => '4910',
'name' => 'The transcriptional regulatory network modulating human trophoblast stem cells to extravillous trophoblast differentiation',
'authors' => 'Kim M. et al.',
'description' => '<p><span>During human pregnancy, extravillous trophoblasts play crucial roles in placental invasion into the maternal decidua and spiral artery remodeling. However, regulatory factors and their action mechanisms modulating human extravillous trophoblast specification have been unknown. By analyzing dynamic changes in transcriptome and enhancer profile during human trophoblast stem cell to extravillous trophoblast differentiation, we define stage-specific regulators, including an early-stage transcription factor, TFAP2C, and multiple late-stage transcription factors. Loss-of-function studies confirm the requirement of all transcription factors identified for adequate differentiation, and we reveal that the dynamic changes in the levels of TFAP2C are essential. Notably, TFAP2C pre-occupies the regulatory elements of the inactive extravillous trophoblast-active genes during the early stage of differentiation, and the late-stage transcription factors directly activate extravillous trophoblast-active genes, including themselves as differentiation further progresses, suggesting sequential actions of transcription factors assuring differentiation. Our results reveal stage-specific transcription factors and their inter-connected regulatory mechanisms modulating extravillous trophoblast differentiation, providing a framework for understanding early human placentation and placenta-related complications.</span></p>',
'date' => '2024-02-12',
'pmid' => 'https://www.nature.com/articles/s41467-024-45669-2',
'doi' => 'https://doi.org/10.1038/s41467-024-45669-2',
'modified' => '2024-02-15 10:43:52',
'created' => '2024-02-15 10:43:52',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 19 => array(
'id' => '4928',
'name' => 'DMSO derives Trophectoderm and Clonal Blastoid from Single Human Pluripotent Stem Cell',
'authors' => 'Alsolami S. et al.',
'description' => '<p><span>Human naïve pluripotent stem cells (nPSCs) can differentiate into extra-embryonic trophectoderm (TE), a critical step in the generation of the integrated embryo model termed blastoid. The current paradigm of blastoid generation necessitates the aggregation of dozens of nPSCs treated with multiple small molecule inhibitors, growth factors, or genetic modifications to initiate TE differentiation. The presence of complex crosstalk among pathways and cellular heterogeneity in these models complicates mechanistic study and genetic screens. Here, we show that a single small molecule, dimethyl sulfoxide (DMSO), potently induces TE differentiation in basal medium without pharmacological and genetic perturbations. DMSO enhances blastoid generation and, more importantly, is sufficient for blastoid generation by itself. DMSO blastoids resemble blastocysts in morphology and lineage composition. DMSO induces blastoid formation through PKC signaling and cell cycle regulation. Lastly, DMSO enables single nPSC-derived clonal blastoids, which could facilitate genetic screens for mechanistic understanding of human embryogenesis.</span></p>',
'date' => '2024-01-01',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2023.12.31.573770v1.abstract',
'doi' => 'https://doi.org/10.1101/2023.12.31.573770',
'modified' => '2024-03-27 15:18:04',
'created' => '2024-03-27 15:18:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 20 => array(
'id' => '4887',
'name' => 'In vitro production of cat-restricted Toxoplasma pre-sexual stages',
'authors' => 'Antunes, A.V. et al.',
'description' => '<p><span>Sexual reproduction of </span><i>Toxoplasma gondii</i><span>, confined to the felid gut, remains largely uncharted owing to ethical concerns regarding the use of cats as model organisms. Chromatin modifiers dictate the developmental fate of the parasite during its multistage life cycle, but their targeting to stage-specific cistromes is poorly described</span><sup><a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 1" title="Farhat, D. C. et al. A MORC-driven transcriptional switch controls Toxoplasma developmental trajectories and sexual commitment. Nat. Microbiol. 5, 570–583 (2020)." href="https://www.nature.com/articles/s41586-023-06821-y#ref-CR1" id="ref-link-section-d277698175e527">1</a>,<a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 2" title="Bougdour, A. et al. Drug inhibition of HDAC3 and epigenetic control of differentiation in Apicomplexa parasites. J. Exp. Med. 206, 953–966 (2009)." href="https://www.nature.com/articles/s41586-023-06821-y#ref-CR2" id="ref-link-section-d277698175e530">2</a></sup><span>. Here we found that the transcription factors AP2XII-1 and AP2XI-2 operate during the tachyzoite stage, a hallmark of acute toxoplasmosis, to silence genes necessary for merozoites, a developmental stage critical for subsequent sexual commitment and transmission to the next host, including humans. Their conditional and simultaneous depletion leads to a marked change in the transcriptional program, promoting a full transition from tachyzoites to merozoites. These in vitro-cultured pre-gametes have unique protein markers and undergo typical asexual endopolygenic division cycles. In tachyzoites, AP2XII-1 and AP2XI-2 bind DNA as heterodimers at merozoite promoters and recruit MORC and HDAC3 (ref. </span><sup><a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 1" title="Farhat, D. C. et al. A MORC-driven transcriptional switch controls Toxoplasma developmental trajectories and sexual commitment. Nat. Microbiol. 5, 570–583 (2020)." href="https://www.nature.com/articles/s41586-023-06821-y#ref-CR1" id="ref-link-section-d277698175e534">1</a></sup><span>), thereby limiting chromatin accessibility and transcription. Consequently, the commitment to merogony stems from a profound epigenetic rewiring orchestrated by AP2XII-1 and AP2XI-2. Successful production of merozoites in vitro paves the way for future studies on<span> </span></span><i>Toxoplasma</i><span><span> </span>sexual development without the need for cat infections and holds promise for the development of therapies to prevent parasite transmission.</span></p>',
'date' => '2023-12-13',
'pmid' => 'https://www.nature.com/articles/s41586-023-06821-y',
'doi' => 'https://doi.org/10.1038/s41586-023-06821-y',
'modified' => '2023-12-18 10:40:50',
'created' => '2023-12-18 10:40:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 21 => array(
'id' => '4939',
'name' => 'Vaginal microbes alter epithelial transcriptomic and epigenomic modifications providing insight into the molecular mechanisms for susceptibility to adverse reproductive outcomes',
'authors' => 'Elovitz M. et al.',
'description' => '<p><span>The cervicovaginal microbiome is highly associated with women's health with microbial communities dominated by </span><i>Lactobacillus</i><span><span> </span>spp. being considered optimal. Conversely, a lack of lactobacilli and a high abundance of strict and facultative anaerobes including<span> </span></span><i>Gardnerella vaginalis</i><span>, have been associated with adverse reproductive outcomes. However, the molecular pathways modulated by microbe interactions with the cervicovaginal epithelia remain unclear. Using RNA-sequencing, we characterize the<span> </span></span><i>in vitro</i><span><span> </span>cervicovaginal epithelial transcriptional response to different vaginal bacteria and their culture supernatants. We showed that<span> </span></span><i>G. vaginalis</i><span><span> </span>upregulated genes were associated with an activated innate immune response including anti-microbial peptides and inflammasome pathways, represented by NLRP3-mediated increases in caspase-1, IL-1β and cell death. Cervicovaginal epithelial cells exposed to<span> </span></span><i>L. crispatus</i><span><span> </span>showed limited transcriptomic changes, while exposure to<span> </span></span><i>L. crispatus</i><span><span> </span>culture supernatants resulted in a shift in the epigenomic landscape of cervical epithelial cells. ATAC-sequencing confirmed epigenetic changes with reduced chromatin accessibility. This study reveals new insight into host-microbe interactions in the lower reproductive tract and suggest potential therapeutic strategies leveraging the vaginal microbiome to improve reproductive health.</span></p>',
'date' => '2023-11-16',
'pmid' => 'https://pubmed.ncbi.nlm.nih.gov/38014044/',
'doi' => '10.21203/rs.3.rs-3580132/v1',
'modified' => '2024-06-07 15:47:48',
'created' => '2024-06-07 15:47:48',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 22 => array(
'id' => '4927',
'name' => 'Inflammatory stress-mediated chromatin changes underlie dysfunction in endothelial cells',
'authors' => 'Liu H. et al.',
'description' => '<p><span>Inflammatory stresses underlie endothelial dysfunction and contribute to the development of chronic cardiovascular disorders such as atherosclerosis and vascular fibrosis. The initial transcriptional response of endothelial cells to pro-inflammatory cytokines such as TNF-alpha is well established. However, very few studies uncover the effects of inflammatory stresses on chromatin architecture. We used integrative analysis of ATAC-seq and RNA-seq data to investigate chromatin alterations in human endothelial cells in response to TNF-alpha and febrile-range heat stress exposure. Multi-omics data analysis suggests a correlation between the transcription of stress-related genes and endothelial dysfunction drivers with chromatin regions exhibiting differential accessibility. Moreover, microscopy identified the dynamics in the nuclear organization, specifically, the changes in a subset of heterochromatic nucleoli-associated chromatin domains, the centromeres. Upon inflammatory stress exposure, the centromeres decreased association with nucleoli in a p38-dependent manner and increased the number of transcripts from pericentromeric regions. Overall, we provide two lines of evidence that suggest chromatin alterations in vascular endothelial cells during inflammatory stresses.</span></p>',
'date' => '2023-10-16',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10614786/',
'doi' => '10.1101/2023.10.11.561959',
'modified' => '2024-03-27 15:15:21',
'created' => '2024-03-27 15:15:21',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 23 => array(
'id' => '4880',
'name' => 'Dynamic PRC1-CBX8 stabilizes a porous structure of chromatin condensates',
'authors' => 'Uckelmann M. et al.',
'description' => '<p><span>The compaction of chromatin is a prevalent paradigm in gene repression. Chromatin compaction is commonly thought to repress transcription by restricting chromatin accessibility. However, the spatial organisation and dynamics of chromatin compacted by gene-repressing factors are unknown. Using cryo-electron tomography, we solved the three dimensional structure of chromatin condensed by the Polycomb Repressive Complex 1 (PRC1) in a complex with CBX8. PRC1-condensed chromatin is porous and stabilised through multivalent dynamic interactions of PRC1 with chromatin. Within condensates, PRC1 remains dynamic while maintaining a static chromatin structure. In differentiated mouse embryonic stem cells, CBX8-bound chromatin remains accessible. These findings challenge the idea of rigidly compacted polycomb domains and instead provides a mechanistic framework for dynamic and accessible PRC1-chromatin condensates.</span></p>',
'date' => '2023-05-09',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2023.05.08.539931v1.abstract',
'doi' => 'https://doi.org/10.1101/2023.05.08.539931',
'modified' => '2023-11-10 15:43:37',
'created' => '2023-11-10 15:43:37',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 24 => array(
'id' => '4810',
'name' => 'UMSBP2 is chromatin remodeler that functions in regulation of geneexpression and suppression of antigenic variation in trypanosomes.',
'authors' => 'Soni A. et al.',
'description' => '<p><span>Universal Minicircle Sequence binding proteins (UMSBPs) are CCHC-type zinc-finger proteins that bind the single-stranded G-rich UMS sequence, conserved at the replication origins of minicircles in the kinetoplast DNA, the mitochondrial genome of kinetoplastids. Trypanosoma brucei UMSBP2 has been recently shown to colocalize with telomeres and to play an essential role in chromosome end protection. Here we report that TbUMSBP2 decondenses in vitro DNA molecules, which were condensed by core histones H2B, H4 or linker histone H1. DNA decondensation is mediated via protein-protein interactions between TbUMSBP2 and these histones, independently of its previously described DNA binding activity. Silencing of the TbUMSBP2 gene resulted in a significant decrease in the disassembly of nucleosomes in T. brucei chromatin, a phenotype that could be reverted, by supplementing the knockdown cells with TbUMSBP2. Transcriptome analysis revealed that silencing of TbUMSBP2 affects the expression of multiple genes in T. brucei, with a most significant effect on the upregulation of the subtelomeric variant surface glycoproteins (VSG) genes, which mediate the antigenic variation in African trypanosomes. These observations suggest that UMSBP2 is a chromatin remodeling protein that functions in the regulation of gene expression and plays a role in the control of antigenic variation in T. brucei.</span></p>',
'date' => '2023-05-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/37207337',
'doi' => '10.1093/nar/gkad402',
'modified' => '2023-06-15 08:54:17',
'created' => '2023-06-13 21:11:31',
'ProductsPublication' => array(
[maximum depth reached]
)
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
'id' => '2440',
'name' => 'ATAC-seq kit SDS US en',
'language' => 'en',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-US-en-1_0.pdf',
'countries' => 'US',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '2438',
'name' => 'ATAC-seq kit SDS GB en',
'language' => 'en',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-GB-en-1_0.pdf',
'countries' => 'GB',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '2437',
'name' => 'ATAC-seq kit SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-FR-fr-1_0.pdf',
'countries' => 'FR',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '2433',
'name' => 'ATAC-seq kit SDS BE fr',
'language' => 'fr',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-BE-fr-1_0.pdf',
'countries' => 'BE',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '2436',
'name' => 'ATAC-seq kit SDS ES es',
'language' => 'es',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-ES-es-1_0.pdf',
'countries' => 'ES',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '2439',
'name' => 'ATAC-seq kit SDS JP ja',
'language' => 'ja',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-JP-ja-1_0.pdf',
'countries' => 'JP',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '2435',
'name' => 'ATAC-seq kit SDS DE de',
'language' => 'de',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-DE-de-1_0.pdf',
'countries' => 'DE',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '2434',
'name' => 'ATAC-seq kit SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-BE-nl-1_0.pdf',
'countries' => 'BE',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
)
)
)
$meta_canonical = 'https://dev.diagenode.com/en/p/atac-seq-kit-24rxns'
$fromEmail = 'dev.diagenode.com'
$country = 'US'
$countries_allowed = array(
(int) 0 => 'CA',
(int) 1 => 'US',
(int) 2 => 'IE',
(int) 3 => 'GB',
(int) 4 => 'DK',
(int) 5 => 'NO',
(int) 6 => 'SE',
(int) 7 => 'FI',
(int) 8 => 'NL',
(int) 9 => 'BE',
(int) 10 => 'LU',
(int) 11 => 'FR',
(int) 12 => 'DE',
(int) 13 => 'CH',
(int) 14 => 'AT',
(int) 15 => 'ES',
(int) 16 => 'IT',
(int) 17 => 'PT'
)
$outsource = false
$other_formats = array(
(int) 0 => array(
'id' => '3193',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<p><a href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p><strong>ATAC-seq</strong>, Assay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the <strong>transposase Tn5</strong> which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
<p>The Diagenode’s <b>ATAC-</b><b>seq</b><b> kit </b>is based on a highly validated protocol optimized for <b>50,000 </b><b>cells</b><b> per </b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The primer indexes for multiplexing are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell </b><b>requirement</b><b>: </b><b>50,000 </b><b>cells / </b><b>rxn</b></li>
<li><b>Robust protocol </b>with <b>high reproducibility </b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong> and <b>efficient DNA capture </b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids </b><b>over-amplification </b></li>
<li>Allows adaptation/flexibility for <b>more challenging samples </b>to succeed with library prep.</li>
<li>Gives <strong>early indication</strong> if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p><span>Looking for ATAC-seq on tissue? Please, go to: </span><a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack: <a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"> 0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"> (Tn5 transposase) </a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"> Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '8 rxns',
'catalog_number' => 'C01080001',
'old_catalog_number' => '',
'sf_code' => 'C01080001-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '615',
'price_USD' => '615',
'price_GBP' => '535',
'price_JPY' => '110700',
'price_CNY' => '',
'price_AUD' => '1538',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-8rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment (C01080001)| Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2023-05-05 07:57:08',
'created' => '2021-06-24 16:47:47',
'ProductsGroup' => array(
'id' => '397',
'product_id' => '3193',
'group_id' => '349'
)
)
)
$pro = array(
'id' => '3193',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<p><a href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p><strong>ATAC-seq</strong>, Assay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the <strong>transposase Tn5</strong> which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
<p>The Diagenode’s <b>ATAC-</b><b>seq</b><b> kit </b>is based on a highly validated protocol optimized for <b>50,000 </b><b>cells</b><b> per </b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The primer indexes for multiplexing are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell </b><b>requirement</b><b>: </b><b>50,000 </b><b>cells / </b><b>rxn</b></li>
<li><b>Robust protocol </b>with <b>high reproducibility </b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong> and <b>efficient DNA capture </b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids </b><b>over-amplification </b></li>
<li>Allows adaptation/flexibility for <b>more challenging samples </b>to succeed with library prep.</li>
<li>Gives <strong>early indication</strong> if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p><span>Looking for ATAC-seq on tissue? Please, go to: </span><a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack: <a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"> 0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"> (Tn5 transposase) </a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"> Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '8 rxns',
'catalog_number' => 'C01080001',
'old_catalog_number' => '',
'sf_code' => 'C01080001-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '615',
'price_USD' => '615',
'price_GBP' => '535',
'price_JPY' => '110700',
'price_CNY' => '',
'price_AUD' => '1538',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-8rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment (C01080001)| Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2023-05-05 07:57:08',
'created' => '2021-06-24 16:47:47',
'ProductsGroup' => array(
'id' => '397',
'product_id' => '3193',
'group_id' => '349'
)
)
$edit = ''
$testimonials = ''
$featured_testimonials = ''
$related_products = ''
$rrbs_service = array(
(int) 0 => (int) 1894,
(int) 1 => (int) 1895
)
$chipseq_service = array(
(int) 0 => (int) 2683,
(int) 1 => (int) 1835,
(int) 2 => (int) 1836,
(int) 3 => (int) 2684,
(int) 4 => (int) 1838,
(int) 5 => (int) 1839,
(int) 6 => (int) 1856
)
$labelize = object(Closure) {
}
$old_catalog_number = ''
$country_code = 'US'
$other_format = array(
'id' => '3193',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<p><a href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p><strong>ATAC-seq</strong>, Assay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the <strong>transposase Tn5</strong> which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
<p>The Diagenode’s <b>ATAC-</b><b>seq</b><b> kit </b>is based on a highly validated protocol optimized for <b>50,000 </b><b>cells</b><b> per </b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The primer indexes for multiplexing are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell </b><b>requirement</b><b>: </b><b>50,000 </b><b>cells / </b><b>rxn</b></li>
<li><b>Robust protocol </b>with <b>high reproducibility </b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong> and <b>efficient DNA capture </b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids </b><b>over-amplification </b></li>
<li>Allows adaptation/flexibility for <b>more challenging samples </b>to succeed with library prep.</li>
<li>Gives <strong>early indication</strong> if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p><span>Looking for ATAC-seq on tissue? Please, go to: </span><a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack: <a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"> 0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"> (Tn5 transposase) </a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"> Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '8 rxns',
'catalog_number' => 'C01080001',
'old_catalog_number' => '',
'sf_code' => 'C01080001-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '615',
'price_USD' => '615',
'price_GBP' => '535',
'price_JPY' => '110700',
'price_CNY' => '',
'price_AUD' => '1538',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-8rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment (C01080001)| Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2023-05-05 07:57:08',
'created' => '2021-06-24 16:47:47',
'ProductsGroup' => array(
'id' => '397',
'product_id' => '3193',
'group_id' => '349'
)
)
$label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>'
$document = array(
'id' => '1139',
'name' => 'ATAC-seq Kit - Manual',
'description' => '<p class="p1">Gene expression is carefully regulated in the cells in order to manage a wide range of biological functions. The structure of chromatin is quite dynamic and contributes to this crucial regulatory process.</p>
<p class="p1">ATAC-seq, Assay for Transposase-Accessible Chromatin, followed by nextgeneration sequencing, is a key technology to easily identify the “open” regions of the chromatin, which are usually associated with permissive gene expression. Indeed, the nuclei of the samples are incubated with a transposase, and only the genomic regions associated with open chromatin will be accessible to this transposase. During the process those regions will be cut and sequencing adaptors will be added, allowing their sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only., giving a map of the chromatin status in the whole genome of the sample.</p>
<p class="p1">The Diagenode’s ATAC-seq kit is based on a highly validated protocol, used for years in our Epigenomics Profiling Services offer and takes advantage of many successful Diagenode’s tools, such as the loaded Tagmentase (Tn5 transposase), the MicroChIP DiaPure Columns and the Primer indexes for tagmented libraries kits.</p>',
'image_id' => null,
'type' => 'Manual',
'url' => 'files/products/kits/atacseq-kit-manual.pdf',
'slug' => 'atacseq-kit-manual',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-06-25 09:51:59',
'created' => '2021-06-25 09:51:59',
'ProductsDocument' => array(
'id' => '3130',
'product_id' => '3194',
'document_id' => '1139'
)
)
$sds = array(
'id' => '2434',
'name' => 'ATAC-seq kit SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-BE-nl-1_0.pdf',
'countries' => 'BE',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
'id' => '4140',
'product_id' => '3194',
'safety_sheet_id' => '2434'
)
)
$publication = array(
'id' => '4810',
'name' => 'UMSBP2 is chromatin remodeler that functions in regulation of geneexpression and suppression of antigenic variation in trypanosomes.',
'authors' => 'Soni A. et al.',
'description' => '<p><span>Universal Minicircle Sequence binding proteins (UMSBPs) are CCHC-type zinc-finger proteins that bind the single-stranded G-rich UMS sequence, conserved at the replication origins of minicircles in the kinetoplast DNA, the mitochondrial genome of kinetoplastids. Trypanosoma brucei UMSBP2 has been recently shown to colocalize with telomeres and to play an essential role in chromosome end protection. Here we report that TbUMSBP2 decondenses in vitro DNA molecules, which were condensed by core histones H2B, H4 or linker histone H1. DNA decondensation is mediated via protein-protein interactions between TbUMSBP2 and these histones, independently of its previously described DNA binding activity. Silencing of the TbUMSBP2 gene resulted in a significant decrease in the disassembly of nucleosomes in T. brucei chromatin, a phenotype that could be reverted, by supplementing the knockdown cells with TbUMSBP2. Transcriptome analysis revealed that silencing of TbUMSBP2 affects the expression of multiple genes in T. brucei, with a most significant effect on the upregulation of the subtelomeric variant surface glycoproteins (VSG) genes, which mediate the antigenic variation in African trypanosomes. These observations suggest that UMSBP2 is a chromatin remodeling protein that functions in the regulation of gene expression and plays a role in the control of antigenic variation in T. brucei.</span></p>',
'date' => '2023-05-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/37207337',
'doi' => '10.1093/nar/gkad402',
'modified' => '2023-06-15 08:54:17',
'created' => '2023-06-13 21:11:31',
'ProductsPublication' => array(
'id' => '6971',
'product_id' => '3194',
'publication_id' => '4810'
)
)
$externalLink = ' <a href="https://www.ncbi.nlm.nih.gov/pubmed/37207337" target="_blank"><i class="fa fa-external-link"></i></a>'CustomErrorHandler::handleError() - APP/Lib/CustomErrorHandler.php, line 9
include - APP/View/Products/view.ctp, line 755
View::_evaluate() - CORE/Cake/View/View.php, line 971
View::_render() - CORE/Cake/View/View.php, line 933
View::render() - CORE/Cake/View/View.php, line 473
Controller::render() - CORE/Cake/Controller/Controller.php, line 963
ProductsController::slug() - APP/Controller/ProductsController.php, line 1055
ReflectionMethod::invokeArgs() - [internal], line ??
Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491
Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193
Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167
[main] - APP/webroot/index.php, line 118
(Notice): Undefined variable: header [APP/View/Products/view.ctp, line 755]Code Context $errorType = self::getErrorType($code);
self::logToDatabase( $errorType, $description, $file, $line);
return parent::handleError( $errorType, $description, $file, $line, $context);
$viewFile = '/var/www/dev.diagenode.com/app/View/Products/view.ctp'
$dataForView = array(
'language' => 'en',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'meta_title' => 'ATAC-seq kit for open chromatin assessment C01080001 | Diagenode ',
'product' => array(
'Product' => array(
'id' => '3194',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<div class="row">
<div class="small-12 medium-8 large-8 columns">
<p><a class="tiny details button" href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf">Download the Manual</a></p>
<p><strong>ATAC-seq</strong>, Assay for<span> </span><strong>T</strong>ransposase-<strong>A</strong>ccessible<span> </span><strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the<span> </span><strong>transposase Tn5</strong><span> </span>which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
</div>
<div class="small-12 medium-4 large-4 columns"><center>
<script>// <![CDATA[
var date = new Date(); var heure = date.getHours(); var jour = date.getDay(); var semaine = Math.floor(date.getDate() / 7) + 1; if (jour === 2 && ( (heure >= 9 && heure < 9.5) || (heure >= 18 && heure < 18.5) )) { document.write('<a href="https://us02web.zoom.us/j/85467619762"><img src="https://www.diagenode.com/img/epicafe-ON.gif"></a>'); } else { document.write('<a href="https://go.diagenode.com/l/928883/2023-04-26/3kq1v"><img src="https://www.diagenode.com/img/epicafe-OFF.png"></a>'); }
// ]]></script>
</center></div>
</div>
<p>Diagenode’s<span> </span><b>ATAC-</b><b>seq</b><b><span> </span>kit<span> </span></b>is based on a highly validated protocol optimized for<span> </span><b>50,000<span> </span></b><b>cells</b><b><span> </span>per<span> </span></b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The <a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">primer indexes for multiplexing</a> are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell<span> </span></b><b>requirement</b><b>:<span> </span></b><b>50,000<span> </span></b><b>cells /<span> </span></b><b>rxn</b></li>
<li><b>Robust protocol<span> </span></b>with<span> </span><b>high reproducibility<span> </span></b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong><span> </span>and<span> </span><b>efficient DNA capture<span> </span></b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids<span> </span></b><b>over-amplification</b></li>
<li>Allows adaptation/flexibility for<span> </span><b>more challenging samples<span> </span></b>to succeed with library prep.</li>
<li>Gives<span> </span><strong>early indication</strong><span> </span>if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p>Looking for ATAC-seq on tissue? Please, go to: <a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack:<span> </span><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"><span> </span>0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"><span> </span>(Tn5 transposase)<span> </span></a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"><span> </span>Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '24 rxns',
'catalog_number' => 'C01080002',
'old_catalog_number' => '',
'sf_code' => 'C01080002-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '1740',
'price_USD' => '1745',
'price_GBP' => '1485',
'price_JPY' => '313200',
'price_CNY' => '',
'price_AUD' => '4362',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-24rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment C01080001 | Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2025-04-17 10:29:58',
'created' => '2021-07-01 15:46:58',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => null,
'name' => null,
'description' => null,
'clonality' => null,
'isotype' => null,
'lot' => null,
'concentration' => null,
'reactivity' => null,
'type' => null,
'purity' => null,
'classification' => null,
'application_table' => null,
'storage_conditions' => null,
'storage_buffer' => null,
'precautions' => null,
'uniprot_acc' => null,
'slug' => null,
'meta_keywords' => null,
'meta_description' => null,
'modified' => null,
'created' => null,
'select_label' => null
),
'Slave' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Group' => array(
'Group' => array(
[maximum depth reached]
),
'Master' => array(
[maximum depth reached]
),
'Product' => array(
[maximum depth reached]
)
),
'Related' => array(),
'Application' => array(),
'Category' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Document' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
),
(int) 8 => array(
[maximum depth reached]
),
(int) 9 => array(
[maximum depth reached]
),
(int) 10 => array(
[maximum depth reached]
),
(int) 11 => array(
[maximum depth reached]
),
(int) 12 => array(
[maximum depth reached]
),
(int) 13 => array(
[maximum depth reached]
),
(int) 14 => array(
[maximum depth reached]
),
(int) 15 => array(
[maximum depth reached]
),
(int) 16 => array(
[maximum depth reached]
),
(int) 17 => array(
[maximum depth reached]
),
(int) 18 => array(
[maximum depth reached]
),
(int) 19 => array(
[maximum depth reached]
),
(int) 20 => array(
[maximum depth reached]
),
(int) 21 => array(
[maximum depth reached]
),
(int) 22 => array(
[maximum depth reached]
),
(int) 23 => array(
[maximum depth reached]
),
(int) 24 => array(
[maximum depth reached]
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
)
)
),
'meta_canonical' => 'https://dev.diagenode.com/en/p/atac-seq-kit-24rxns',
'fromEmail' => 'dev.diagenode.com'
)
$language = 'en'
$meta_keywords = ''
$meta_description = 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility'
$meta_title = 'ATAC-seq kit for open chromatin assessment C01080001 | Diagenode '
$product = array(
'Product' => array(
'id' => '3194',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<div class="row">
<div class="small-12 medium-8 large-8 columns">
<p><a class="tiny details button" href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf">Download the Manual</a></p>
<p><strong>ATAC-seq</strong>, Assay for<span> </span><strong>T</strong>ransposase-<strong>A</strong>ccessible<span> </span><strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the<span> </span><strong>transposase Tn5</strong><span> </span>which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
</div>
<div class="small-12 medium-4 large-4 columns"><center>
<script>// <![CDATA[
var date = new Date(); var heure = date.getHours(); var jour = date.getDay(); var semaine = Math.floor(date.getDate() / 7) + 1; if (jour === 2 && ( (heure >= 9 && heure < 9.5) || (heure >= 18 && heure < 18.5) )) { document.write('<a href="https://us02web.zoom.us/j/85467619762"><img src="https://www.diagenode.com/img/epicafe-ON.gif"></a>'); } else { document.write('<a href="https://go.diagenode.com/l/928883/2023-04-26/3kq1v"><img src="https://www.diagenode.com/img/epicafe-OFF.png"></a>'); }
// ]]></script>
</center></div>
</div>
<p>Diagenode’s<span> </span><b>ATAC-</b><b>seq</b><b><span> </span>kit<span> </span></b>is based on a highly validated protocol optimized for<span> </span><b>50,000<span> </span></b><b>cells</b><b><span> </span>per<span> </span></b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The <a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">primer indexes for multiplexing</a> are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell<span> </span></b><b>requirement</b><b>:<span> </span></b><b>50,000<span> </span></b><b>cells /<span> </span></b><b>rxn</b></li>
<li><b>Robust protocol<span> </span></b>with<span> </span><b>high reproducibility<span> </span></b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong><span> </span>and<span> </span><b>efficient DNA capture<span> </span></b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids<span> </span></b><b>over-amplification</b></li>
<li>Allows adaptation/flexibility for<span> </span><b>more challenging samples<span> </span></b>to succeed with library prep.</li>
<li>Gives<span> </span><strong>early indication</strong><span> </span>if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p>Looking for ATAC-seq on tissue? Please, go to: <a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack:<span> </span><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"><span> </span>0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"><span> </span>(Tn5 transposase)<span> </span></a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"><span> </span>Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '24 rxns',
'catalog_number' => 'C01080002',
'old_catalog_number' => '',
'sf_code' => 'C01080002-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '1740',
'price_USD' => '1745',
'price_GBP' => '1485',
'price_JPY' => '313200',
'price_CNY' => '',
'price_AUD' => '4362',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-24rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment C01080001 | Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2025-04-17 10:29:58',
'created' => '2021-07-01 15:46:58',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => null,
'name' => null,
'description' => null,
'clonality' => null,
'isotype' => null,
'lot' => null,
'concentration' => null,
'reactivity' => null,
'type' => null,
'purity' => null,
'classification' => null,
'application_table' => null,
'storage_conditions' => null,
'storage_buffer' => null,
'precautions' => null,
'uniprot_acc' => null,
'slug' => null,
'meta_keywords' => null,
'meta_description' => null,
'modified' => null,
'created' => null,
'select_label' => null
),
'Slave' => array(
(int) 0 => array(
'id' => '349',
'name' => 'C01080002',
'product_id' => '3194',
'modified' => '2021-07-01 15:47:50',
'created' => '2021-07-01 15:47:50'
)
),
'Group' => array(
'Group' => array(
'id' => '349',
'name' => 'C01080002',
'product_id' => '3194',
'modified' => '2021-07-01 15:47:50',
'created' => '2021-07-01 15:47:50'
),
'Master' => array(
'id' => '3194',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<div class="row">
<div class="small-12 medium-8 large-8 columns">
<p><a class="tiny details button" href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf">Download the Manual</a></p>
<p><strong>ATAC-seq</strong>, Assay for<span> </span><strong>T</strong>ransposase-<strong>A</strong>ccessible<span> </span><strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the<span> </span><strong>transposase Tn5</strong><span> </span>which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
</div>
<div class="small-12 medium-4 large-4 columns"><center>
<script>// <![CDATA[
var date = new Date(); var heure = date.getHours(); var jour = date.getDay(); var semaine = Math.floor(date.getDate() / 7) + 1; if (jour === 2 && ( (heure >= 9 && heure < 9.5) || (heure >= 18 && heure < 18.5) )) { document.write('<a href="https://us02web.zoom.us/j/85467619762"><img src="https://www.diagenode.com/img/epicafe-ON.gif"></a>'); } else { document.write('<a href="https://go.diagenode.com/l/928883/2023-04-26/3kq1v"><img src="https://www.diagenode.com/img/epicafe-OFF.png"></a>'); }
// ]]></script>
</center></div>
</div>
<p>Diagenode’s<span> </span><b>ATAC-</b><b>seq</b><b><span> </span>kit<span> </span></b>is based on a highly validated protocol optimized for<span> </span><b>50,000<span> </span></b><b>cells</b><b><span> </span>per<span> </span></b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The <a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">primer indexes for multiplexing</a> are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell<span> </span></b><b>requirement</b><b>:<span> </span></b><b>50,000<span> </span></b><b>cells /<span> </span></b><b>rxn</b></li>
<li><b>Robust protocol<span> </span></b>with<span> </span><b>high reproducibility<span> </span></b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong><span> </span>and<span> </span><b>efficient DNA capture<span> </span></b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids<span> </span></b><b>over-amplification</b></li>
<li>Allows adaptation/flexibility for<span> </span><b>more challenging samples<span> </span></b>to succeed with library prep.</li>
<li>Gives<span> </span><strong>early indication</strong><span> </span>if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p>Looking for ATAC-seq on tissue? Please, go to: <a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack:<span> </span><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"><span> </span>0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"><span> </span>(Tn5 transposase)<span> </span></a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"><span> </span>Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '24 rxns',
'catalog_number' => 'C01080002',
'old_catalog_number' => '',
'sf_code' => 'C01080002-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '1740',
'price_USD' => '1745',
'price_GBP' => '1485',
'price_JPY' => '313200',
'price_CNY' => '',
'price_AUD' => '4362',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-24rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment C01080001 | Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2025-04-17 10:29:58',
'created' => '2021-07-01 15:46:58'
),
'Product' => array(
(int) 0 => array(
[maximum depth reached]
)
)
),
'Related' => array(),
'Application' => array(),
'Category' => array(
(int) 0 => array(
'id' => '13',
'position' => '2',
'parent_id' => null,
'name' => 'Chromatin studies',
'description' => '<div class="row">
<div class="small-12 medium-8 large-8 columns"><br />
<p>Chromatin structure plays a key role in regulating gene expression by allowing DNA accessibility to transcriptional machinery and transcription factors. The packaging of DNA into nucleosomes forms a closed structure that is not highly accessible to transcriptional elements whereas the open nucleosome structure allows DNA to be accessible. Diagenode offers a number of solutions to help you analyze chromatin and the role of transcriptional machinery including ChIP kits, ChIPmentation kits, antibodies, pA-Tn5 and ATAC-seq kits.</p>
</div>
<div class="small-12 medium-4 large-4 columns"><center>
<script>// <![CDATA[
var date = new Date(); var heure = date.getHours(); var jour = date.getDay(); var semaine = Math.floor(date.getDate() / 7) + 1; if (jour === 2 && ( (heure >= 9 && heure < 9.5) || (heure >= 18 && heure < 18.5) )) { document.write('<a href="https://us02web.zoom.us/j/85467619762"><img src="https://www.diagenode.com/img/epicafe-ON.gif"></a>'); } else { document.write('<a href="https://go.diagenode.com/l/928883/2023-04-26/3kq1v"><img src="https://www.diagenode.com/img/epicafe-OFF.png"></a>'); }
// ]]></script>
</center></div>
</div>
<p><a href="https://www.diagenode.com/en/p/uchipmentation-for-histones-24-rxns"><img src="https://www.diagenode.com/img/banners/b-microchip-category.png" /></a></p>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<div id="portal" class="main-portal">
<div class="portal-inner"><nav class="portal-nav">
<ul class="tips-menu">
<li><a href="#workflow" class="tips portal button" style="background: #13b29c; color: #f3fbfa;">Chromatin immunoprecipitation</a></li>
<li><a href="https://www.diagenode.com/en/categories/chromatin-ip-chipmentation" class="tips portal button">ChIPmentation</a></li>
<li><a href="https://www.diagenode.com/en/categories/antibodies
" class="tips portal button">Antibodies</a></li>
<li><a href="https://www.diagenode.com/en/categories/cutandtag" class="tips portal button">pA-Tn5</a></li>
<li><a href="https://www.diagenode.com/en/categories/atac-seq" class="tips portal button">ATAC-seq</a></li>
<li><a href="https://www.diagenode.com/en/categories/cutandtag" class="tips portal button">CUT&Tag</a></li>
</ul>
</nav></div>
</div>
<p>Chromatin immunoprecipitation (ChIP) determines the location of DNA binding sites on the genome for a protein of interest, giving insights into gene expression regulation. ChIP involves the selective enrichment of a chromatin fraction containing a specific antigen. Antibodies that recognize a protein or protein modification are used to determine the relative abundance of that antigen at a specific locus or loci. ChIP can be used to compare enrichment of proteins, map protein modifications, or quantify a protein modification during a time course.</p>
<span class="anchor" id="workflow"></span>
<h4><span style="font-weight: 400;">The ChIP workflow</span></h4>
<ul class="accordion" data-accordion="">
<li class="accordion-navigation"><img src="https://www.diagenode.com/img/categories/kits_chromatin_function/website-chip-workflow.jpg" />
<div id="chip_workflow" class="content">
<div class="row">
<table>
<tbody>
<tr>
<td width="50%">
<h3 class="text-center">Step by step workflow</h3>
</td>
<td width="50%">
<h3 class="text-center">Optimal solution from Diagenode</h3>
</td>
</tr>
<tr>
<td colspan="2"></td>
</tr>
<tr valign="top">
<td>
<p class="lead text-center"><strong>1.</strong><span> </span>Crosslink to bind proteins to DNA</p>
<center><img src="https://www.diagenode.com/img/chip/step-by-step-1-workflow.png" width="192" height="24" /></center></td>
<td>
<ul class="arrow">
<li><a href="https://www.diagenode.com/p/chip-cross-link-gold-600-ul">ChIP Cross-link Gold</a></li>
</ul>
</td>
</tr>
<tr>
<td colspan="2"></td>
</tr>
<tr valign="top">
<td>
<p class="lead text-center"><strong>2.</strong><span> </span>Shear DNA</p>
<center><img src="https://www.diagenode.com/img/chip/step-by-step-2-workflow.png" width="194" height="80" /></center></td>
<td>
<ul class="arrow">
<li><a href="https://www.diagenode.com/p/bioruptor-pico-sonication-device#">Bioruptor<sup>®</sup><span> </span>Pico Sonication device</a></li>
<li><a href="https://www.diagenode.com/categories/chromatin-shearing">Shearing optimization reagent</a></li>
</ul>
</td>
</tr>
<tr>
<td colspan="2"></td>
</tr>
<tr valign="top">
<td>
<p class="lead text-center"><strong>3.</strong><span> </span>Immunoprecipitate with specific antibody</p>
<center><img src="https://www.diagenode.com/img/chip/step-by-step-3-workflow.png" width="47" height="51" /></center></td>
<td>
<ul class="arrow">
<li><a href="https://www.diagenode.com/categories/chromatin-immunoprecipitation">ChIP-seq and ChIP-qPCR kits</a><span> </span>for transcription factors, histones, low inputs, plants</li>
<li><a href="https://www.diagenode.com/categories/chip-grade-antibodies">ChIP</a><span> </span>and<span> </span><a href="https://www.diagenode.com/categories/chip-seq-grade-antibodies">ChIP-seq grade</a><span> </span>antibodies</li>
<li><a href="https://www.diagenode.com/categories/ip-star">Automation available</a></li>
</ul>
</td>
</tr>
<tr>
<td colspan="2"></td>
</tr>
<tr valign="top">
<td>
<p class="lead text-center"><strong>4.</strong><span> </span>Reverse crosslinks and purify</p>
<center><img src="https://www.diagenode.com/img/chip/step-by-step-4-workflow.png" width="194" height="80" /></center></td>
<td>
<ul class="arrow">
<li>Diagenode's ChIP kits (contain optimal purification modules)</li>
</ul>
</td>
</tr>
<tr>
<td colspan="2"></td>
</tr>
<tr valign="top">
<td>
<p class="lead text-center"><strong>5.</strong><span> </span><a href="https://www.diagenode.com/en/p/ideal-chip-qpcr-kit">ChIP-qPCR</a><span> </span>or ChIP-seq library preparation</p>
</td>
<td>
<ul class="arrow">
<li>ChIP-seq:</li>
</ul>
<ul style="list-style-type: square;">
<li style="margin-left: 40px; font-size: 1.09rem;"><a href="https://www.diagenode.com/p/microplex-library-preparation-kit-v2-x48-12-indices-48-rxns">MicroPlex Library Preparation for 50 pg - 5 ng</a></li>
<li style="margin-left: 40px; font-size: 1.09rem;"><a href="https://www.diagenode.com/p/ideal-library-preparation-kit-x24-incl-index-primer-set-1-24-rxns">iDeal Library Preparation for > 5 ng</a></li>
</ul>
<ul class="arrow">
<li>ChIP qPCR</li>
</ul>
</td>
</tr>
</tbody>
</table>
</div>
</div>
</li>
</ul>
<p></p>
<h4><span style="font-weight: 400;">Products for chromatin study</span></h4>
<p><span style="font-weight: 400;"></span></p>
<p style="padding-left: 30px;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/crosslinking.png" alt="" width="35" height="35" /> Crosslinking<br /></b></span></strong><span style="font-weight: 400;">Efficient solution for protein-protein fixation.</span><strong><span style="font-weight: 400;"><b><span style="font-weight: 400;"> <a href="../p/chip-cross-link-gold-600-ul
">Read more</a></span></b><br /></span></strong></p>
<p style="padding-left: 30px;"><strong><img src="https://www.diagenode.com/img/applications/chromatin-shearing.png" alt="" width="35" height="35" /> Chromatin shearing<br /></strong><strong><span style="font-weight: 400;">Perfectly sheared chromatin is critical for ChIP success.<span> </span><br /></span></strong><strong><a href="../categories/chromatin-shearing"><span style="font-weight: 400;">Read more</span></a><span style="font-weight: 400;"><span> </span>about solutions for successful chromatin preparation.<span> </span><br /></span></strong><strong><span style="font-weight: 400;"><a href="../categories/bioruptor-shearing-device">Read more</a><span> </span>about<span> </span></span><span style="font-weight: 400;">Bioruptor<span> </span></span><span style="font-weight: 400;">sonication.</span></strong></p>
<p style="padding-left: 30px;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/immunoprecipitation.png" width="35" height="35" caption="false" /> Chromatin immunoprecipitation<br /></b></span></strong><span style="font-weight: 400;">Immunoprecipitation solutions for histone and transcription factor ChIP-qPCR and ChIP-seq for low inputs, plants, and animals including automated solutions</span><strong><span style="font-weight: 400;"><b><span style="font-weight: 400;">. <a href="../categories/chromatin-immunoprecipitation">Read more</a></span></b><br /></span></strong></p>
<p style="padding-left: 30px;"><strong><img src="https://www.diagenode.com/img/applications/ChIPmentation.png" alt="" width="35" height="35" /> ChIPmentation<br /><span style="font-weight: 400;">ChIPmentation, an exclusive technology, is an end-to-end ChIP-seq solution for low and difficult inputs. <a href="../categories/chromatin-ip-chipmentation">Read more</a></span><br /></strong></p>
<p style="padding-left: 30px;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/ChIP-antibodies.png" alt="" width="35" height="35" /> ChIP and ChIP-seq antibodies<br /></b></span></strong><span style="font-weight: 400;">ChIP-grade antibodies are essential for success. <a href="../categories/antibodies">Learn more</a></span><span style="font-weight: 400;"><br /></span></p>
<p style="padding-left: 30px;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/ChIP-qPCR.png" width="35" height="35" caption="false" /> Primer pairs<br /></b></span></strong><span style="font-weight: 400;">Highly specific primer pairs for the amplification of the specific genomic regions. <a href="../categories/primer-pairs">Read more</a></span><span style="font-weight: 400;"><br /></span></p>
<p style="padding-left: 30px;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/cutandtag.png" width="35" height="35" caption="false" /> CUT&Tag solutions<br /></b></span></strong><span style="font-weight: 400;">An alternative to ChIP-seq that combines antibody-targeted controlled cleavage by a protein A-Tn5 fusion with NGS to identify the binding sites of DNA-associated proteins. <a href="../categories/cutandtag">Read more</a></span><span style="font-weight: 400;"><br /></span></p>
<p style="padding-left: 30px;"><span style="font-weight: 400;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/dna-purification.png" width="35" height="35" caption="false" /> DNA purification<br /></b></span></strong><span style="font-weight: 400;"><a href="../categories/dna-and-rna-purification">Read more</a> about solutions for DNA purification</span></span></p>
<p style="padding-left: 30px;"><span style="font-weight: 400;"><span style="font-weight: 400;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/ChIP-seq.png" width="35" height="35" caption="false" /> Library preparation for ChIP-seq<br /></b></span></strong><span style="font-weight: 400;">Optimized solutions for the library preparation from low DNA input. <a href="../categories/library-preparation-for-ChIP-seq">Read more</a></span></span></span></p>
<p style="padding-left: 30px;"><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/ChIP-automation.png" alt="" width="35" height="35" /> ChIP and ChIP-seq automation<br /></b>Reproducibility, optimization simplicity, no variability. <a href="../categories/epigenetic-automation">Learn more</a></span></p>
<div class="row" style="margin-top: 32px;">
<div class="small-12 medium-10 large-9 small-centered columns">
<div class="radius panel" style="background-color: #fff;">
<p class="text-justify">We offer <a href="https://www.diagenode.com/en/categories/chromatin-immunoprecipitation" target="_blank">complete ChIP kits</a> or <strong>individual kit components</strong> from antibodies, buffers, beads, chromatin shearing, and purification reagents. With the ChIP Kit Customizer, you have complete flexibility on which components you want from our validated ChIP kits.</p>
<div class="center" style="text-align: center;"><a href="../pages/chip-kit-customizer-1"><img src="https://www.diagenode.com/img/banners/banner-customizer.png" alt="" /></a></div>
</div>
</div>
</div>
<h4>Chromatin resources</h4>
<h3>Posters</h3>
<ul>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/antibody-chipseq-qc-using-the-ipstar-compact-poster"><span style="font-weight: 400;">Understanding our antibody QC</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/antibodies-you-can-trust-poster"><span style="font-weight: 400;">High quality ChIP antibodies</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/chip-kit-results-with-true-microchip-kit-poster"><span style="font-weight: 400;">ChIP with only 10,000 cells</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/high-resolution-chipseq-profiles-with-ipstar-automated-platform-poster"><span style="font-weight: 400;">High resolution ChIP-seq using automation</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/bioinformatics-pipeline-for-chipseq-analyses"><span style="font-weight: 400;">ChIP-seq bioinformatics</span></a></li>
</ul>
<h3><span style="font-weight: 400;">Application notes</span></h3>
<ul>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/chipettor-application-note"><span style="font-weight: 400;">Simple semi-automaton for easy and inexpensive ChIP</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/chipseq-from-human-tumor-tissue"><span style="font-weight: 400;">Performing ChIP-seq on human tumor tissue</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/plant-chip-seq-application-note"><span style="font-weight: 400;">Plant ChIP-seq – a successful method</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/chip-seq-application-note"><span style="font-weight: 400;">Best workflow practices for low input ChIP</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/files/application_notes/AN-ChIP-Cas9-02_2018.pdf"><span style="font-weight: 400;">Optimize the selection of guide RNA by ChIP to keep CRISPR on-target</span></a></li>
</ul>
<h3>Publications related to ChIP</h3>
<ul>
<li><a href="https://www.diagenode.com/en/publications/view/3373">Corticosteroid receptors adopt distinct cyclical transcriptional signatures</a></li>
<li><a href="https://www.diagenode.com/en/publications/view/3347">Pro-inflammatory cytokine and high doses of ionizing radiation have similar effects on the expression of NF-kappaB-dependent genes</a></li>
<li><a href="https://www.diagenode.com/en/publications/view/3355">Functional dissection of Drosophila melanogaster SUUR protein influence on H3K27me3 profile</a></li>
</ul>
<h3><span style="font-weight: 400;">Brochures</span></h3>
<ul>
<li><a href="https://www.diagenode.com/files/brochures/Chromatin_Immunoprecipitation_Brochure.pdf"><b>Chromatin </b><span style="font-weight: 400;">products brochure</span></a></li>
<li><a href="https://www.diagenode.com/files/brochures/Epigenetic_Antibodies_Brochure.pdf"><span style="font-weight: 400;">Epigenetic<span> </span></span><b>Antibodies</b></a></li>
<li><a href="https://www.diagenode.com/files/brochures/Bioruptor_Sonicator_Brochure.pdf"><span style="font-weight: 400;">Bioruptor for<span> </span></span><b>chromatin shearing</b></a></li>
<li><a href="https://www.diagenode.com/files/brochures/IPStar_Automated_System_Brochure.pdf"><span style="font-weight: 400;">Automating<span> </span></span><b>ChIP</b><span style="font-weight: 400;"><span> </span>and<span> </span></span><b>ChIP-seq</b></a></li>
</ul>
</div>
</div>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => true,
'hide' => false,
'all_format' => false,
'is_antibody' => false,
'slug' => 'chromatin-function',
'cookies_tag_id' => null,
'meta_keywords' => 'Chromatin function,Chromatin shearing,Chromatin immunoprecipitation,Chromatin assembly',
'meta_description' => 'Diagenode offers a number of unique solutions and kits to make Diagenode offers chromatin immunoprecipitation kits have been designed to meet a wide variety of research needs and accessible',
'meta_title' => 'Chromatin Function Kit for Unique Solutions and Kits to make Chromatin Research| Diagenode',
'modified' => '2025-05-21 12:06:52',
'created' => '2015-05-27 05:22:15',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
)
),
'Document' => array(
(int) 0 => array(
'id' => '1139',
'name' => 'ATAC-seq Kit - Manual',
'description' => '<p class="p1">Gene expression is carefully regulated in the cells in order to manage a wide range of biological functions. The structure of chromatin is quite dynamic and contributes to this crucial regulatory process.</p>
<p class="p1">ATAC-seq, Assay for Transposase-Accessible Chromatin, followed by nextgeneration sequencing, is a key technology to easily identify the “open” regions of the chromatin, which are usually associated with permissive gene expression. Indeed, the nuclei of the samples are incubated with a transposase, and only the genomic regions associated with open chromatin will be accessible to this transposase. During the process those regions will be cut and sequencing adaptors will be added, allowing their sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only., giving a map of the chromatin status in the whole genome of the sample.</p>
<p class="p1">The Diagenode’s ATAC-seq kit is based on a highly validated protocol, used for years in our Epigenomics Profiling Services offer and takes advantage of many successful Diagenode’s tools, such as the loaded Tagmentase (Tn5 transposase), the MicroChIP DiaPure Columns and the Primer indexes for tagmented libraries kits.</p>',
'image_id' => null,
'type' => 'Manual',
'url' => 'files/products/kits/atacseq-kit-manual.pdf',
'slug' => 'atacseq-kit-manual',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-06-25 09:51:59',
'created' => '2021-06-25 09:51:59',
'ProductsDocument' => array(
[maximum depth reached]
)
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
'id' => '1819',
'name' => 'product/kits/atacseq-kit-picture.jpg',
'alt' => 'ATAC-seq kit',
'modified' => '2021-06-25 10:03:22',
'created' => '2021-06-25 10:03:22',
'ProductsImage' => array(
[maximum depth reached]
)
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
'id' => '5143',
'name' => 'CRAMP1 drives linker histone expression to enable Polycomb repression',
'authors' => 'Matthews, Rachael E et al.',
'description' => '<div class="abstract-content selected" id="eng-abstract">
<p style="text-align: justify;">In contrast to the well-understood role of core histones in DNA packaging, the function of the linker histone (H1) remains enigmatic. Challenging the prevailing view that linker histones are a general feature of heterochromatin, here we show a critical requirement for H1 in Polycomb repressive complex 2 (PRC2) function. A CRISPR-Cas9 genetic screen using a fluorescent PRC2 reporter identified an essential role for the poorly characterized gene CRAMP1 in PRC2-mediated repression. CRAMP1 localizes to the promoters of expressed H1 genes and positively regulates their transcription. CRAMP1 ablation simultaneously depletes all linker histones, which results in selective decompaction of H3K27me3-marked loci and derepression of PRC2 target genes without concomitant loss of PRC2 occupancy or enzymatic activity. Strikingly, we find that linker histones preferentially localize to genomic loci marked by H3K27me3 across diverse cell types and organisms. Altogether, these data demonstrate a prominent role for linker histones in epigenetic repression by PRC2.</p>
</div>',
'date' => '2025-06-10',
'pmid' => 'https://pubmed.ncbi.nlm.nih.gov/40516528/',
'doi' => '10.1016/j.molcel.2025.05.031',
'modified' => '2025-06-18 09:45:01',
'created' => '2025-06-18 09:45:01',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '5142',
'name' => 'Temporal and spatial niche partitioning in a retrotransposon community of the Drosophila melanogaster genome',
'authors' => 'Varoqui, Marion et al.',
'description' => '<div class="abstract" id="abstract">
<div class="abstract-content selected" id="eng-abstract">
<p style="text-align: justify;">Transposable elements (TEs) are genetic parasites that can potentially threaten the stability of the genomes they colonize. Nonetheless, TEs persist within genomes and are rarely fully eliminated, with diverse TE families coexisting in varing copy numbers. The TE replication strategies that enable host organisms to tolerate and accommodate the extensive diversity of TEs, while minimizing harm to the host and avoiding mutual competition among TEs, remain poorly understood. Here, by studying the spontaneous or experimental mobilization of four Drosophila LTR RetroTransposable Elements (LTR-RTEs), we reveal that each of them preferentially targets open chromatin regions characterized by specific epigenetic features. Among these, gtwin and ZAM are expressed in distinct cell types within female somatic gonadal tissues and inserted into the distinct accessible chromatin landscapes of the corresponding stages of embryogenesis. These findings suggest that individual LTR-RTEs exploit unique biological niches, enabling their coexistence within the tightly regulated ecosystem of the same host genome.</p>
</div>
</div>',
'date' => '2025-06-06',
'pmid' => 'https://pubmed.ncbi.nlm.nih.gov/40503684/',
'doi' => '10.1093/nar/gkaf516',
'modified' => '2025-06-18 09:43:08',
'created' => '2025-06-18 09:43:08',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '5128',
'name' => 'Cervicovaginal microbiome alters transcriptomic and epigenomic signatures across cervicovaginal epithelial barriers',
'authors' => 'Lauren Anton et al. ',
'description' => '<p><b>Background</b></p>
<p>The cervicovaginal microbiome plays a critical role in women's health, with microbial communities dominated by<span> </span><em>Lactobacillus</em><span> </span>species considered optimal. In contrast, the depletion of lactobacilli and the presence of a diverse array of strict and facultative anaerobes, such as<span> </span><em>Gardnerella vaginalis</em>, have been linked with adverse reproductive outcomes. Despite these associations, the molecular mechanisms by which host-microbial interactions modulate cervical and vaginal epithelial function remains poorly understood.</p>
<p><b>Results</b></p>
<p>In this study, we used RNA sequencing to characterize the transcriptional response of cervicovaginal epithelial cells exposed to the culture supernatants of common vaginal bacteria. Our findings revealed that<span> </span><em>G. vaginalis</em><span> </span>culture supernatants upregulate genes associated with an activated innate immune response and increased cell death. Conversely,<span> </span><em>Lactobacillus crispatus</em><span> </span>culture supernatants induced transcriptional changes indicative of epigenomic modeling in ectocervical epithelial cells. Epigenomic modification by<span> </span><em>L. crispatus</em>, was confirmed by ATAC-sequencing, which demonstrated reduced chromatin accessibility.</p>
<p><b>Conclusions</b></p>
<p>These results provide new insights into host-microbe interactions within the lower reproductive tract and suggests that modulating the vaginal microbiome could offer innovative therapeutic strategies to improve reproductive health.</p>',
'date' => '2025-05-07',
'pmid' => 'https://www.researchsquare.com/article/rs-6171614/v1',
'doi' => 'https://doi.org/10.21203/rs.3.rs-6171614/v1',
'modified' => '2025-05-12 10:30:24',
'created' => '2025-05-12 10:30:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '5125',
'name' => 'An ISWI-related chromatin remodeller regulates stage-specific gene expression in Toxoplasma gondii',
'authors' => 'Pachano, Belen et al.',
'description' => '<p style="text-align: justify;"><span>ATP-dependent chromatin remodellers are specialized multiprotein machines that organize the genome in eukaryotic cells and regulate its accessibility by repositioning, ejecting or modifying nucleosomes. However, their role in </span><i>Toxoplasma gondii</i><span><span> </span>is poorly understood. Here we show that<span> </span></span><i>T.</i><span> </span><i>gondii</i><span><span> </span>has evolved two divergent proteins within the imitation switch (ISWI) family:<span> </span></span><i>Tg</i><span>SNF2h and<span> </span></span><i>Tg</i><span>SNF2L.<span> </span></span><i>Tg</i><span>SNF2h specifically forms a core complex with the transcription factor AP2VIII-2 and the scaffold protein<span> </span></span><i>Tg</i><span>RFTS. Depletion of<span> </span></span><i>Tg</i><span>RFTS phenocopies the knockdown of<span> </span></span><i>Tg</i><span>SNF2h, restricting access to chromatin and altering local gene expression. At the genomic level,<span> </span></span><i>Tg</i><span>SNF2h insulates highly transcribed genes from silenced neighbours, ensuring stage-specific gene regulation. By modulating chromatin accessibility to transcription factors,<span> </span></span><i>Tg</i><span>SNF2h exerts epistatic control over MORC, a key regulator of sexual commitment. Our findings show that a specific ISWI complex orchestrates the partitioning of developmental genes and ensures transcriptional fidelity throughout the parasite life cycle.</span></p>',
'date' => '2025-04-11',
'pmid' => 'https://www.nature.com/articles/s41564-025-01980-2#citeas',
'doi' => 'https://doi.org/10.1038/s41564-025-01980-2',
'modified' => '2025-05-19 11:36:47',
'created' => '2025-05-06 15:49:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '5101',
'name' => 'Repression of oxidative phosphorylation by NR2F2, MTERF3 and GDF15 in human skin under high-glucose stress',
'authors' => 'S. Ley-Ngardigal et al.',
'description' => '<p><span>Lifestyle factors such as a Western diet or metabolic diseases like diabetes disrupt glucose homeostasis and induce stress responses, yet their impact on skin metabolism and structural integrity remains poorly understood. Here, we performed multiomic and bioenergetic analyses of human dermal fibroblasts (HDFs), human equivalent dermis (HED), human reconstructed skin (HRS), and skin explants from diabetic patients. We found that 12 mM glucose stress represses oxidative phosphorylation (OXPHOS) through a dual mechanism: the glucose-dependent nuclear receptor NR2F2 activates mitochondrial transcription termination factor 3 (MTERF3) while inhibiting growth-differentiation factor 15 (GDF15). Promoter assays revealed that MTERF3 is regulated by NR2F2 and MYCN, whereas GDF15 is modulated by NR2F2 and FOS. Consequently, OXPHOS proteins and mitochondrial respiration were suppressed, and MTERF3 overexpression additionally interfered with collagen biosynthesis. In contrast, GDF15 supplementation fully rescued hyperglycemia-induced bioenergetic and metabolomic alterations, suggesting a pharmacological strategy to mitigate hyperglycemic damage in the skin. Finally, silencing GDF15 or TFAM impaired fibroblast haptotaxis and skin reconstruction, underscoring the crucial role of mitochondrial energetics in dermal structure and function. Collectively, these findings identify the NR2F2–MTERF3–GDF15 axis as a key mediator of OXPHOS suppression and highlight a potential therapeutic target to preserve skin integrity under hyperglycemic stress.</span></p>',
'date' => '2025-03-27',
'pmid' => 'https://www.sciencedirect.com/science/article/pii/S2213231725001260',
'doi' => 'https://doi.org/10.1016/j.redox.2025.103613',
'modified' => '2025-03-31 13:44:10',
'created' => '2025-03-31 13:44:10',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '5092',
'name' => 'CRISPR screening identifies regulators of enhancer-mediated androgen receptor transcription in advanced prostate cancer',
'authors' => 'Rachel R. Xiang et al. ',
'description' => '<section id="author-highlights-abstract" property="abstract" typeof="Text" role="doc-abstract">
<h2 property="name"></h2>
</section>
<section id="author-abstract" property="abstract" typeof="Text" role="doc-abstract">
<div id="abspara0010" role="paragraph">Amplification of the androgen receptor (AR) locus is the most frequent alteration in metastatic castration-resistant prostate cancer (CRPC). Recently, it was discovered that an enhancer of the AR is co-amplified with the AR gene body and contributes to increased AR transcription and resistance to androgen deprivation therapy. However, the mechanism of enhancer activation in advanced disease is unknown. Here, we used CRISPR-Cas9 screening to identify transcription factors that bind to the AR enhancer and modulate enhancer-mediated AR transcription. We demonstrate that HOXB13, GATA2, and TFAP2C bind the AR enhancer in patient-derived xenografts and directly impact features associated with an active chromatin state. Interestingly, the AR enhancer belongs to a set of regulatory elements that require HOXB13 to maintain FOXA1 binding, further delineating the role of HOXB13 in CRPC. This work provides a framework to functionally identify<span> </span><i>trans</i>-acting factors required for the activation of disease-related noncoding regulatory elements.</div>
</section>',
'date' => '2025-02-14',
'pmid' => 'https://www.cell.com/cell-reports/fulltext/S2211-1247(25)00083-X',
'doi' => '10.1016/j.celrep.2025.115312',
'modified' => '2025-03-27 16:56:52',
'created' => '2025-03-27 16:54:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '5038',
'name' => 'Schistosoma mansoni antigen induced innate immune memory features mitochondrial biogenesis and can be inhibited by ovarian produced hormones',
'authors' => 'Juan Marcos Oviedo et al.',
'description' => '<p><span>We have previously identified that </span><em>S. mansoni</em><span><span> </span>infection induces a unique form of myeloid training that protects male but not female mice from high fat diet induced disease. Here we demonstrate that ovarian derived hormones account for this sex specific difference. Ovariectomy of females prior to infection permits metabolic reprogramming of the myeloid lineage, with BMDM exhibiting carbon source flexibility for cellular respiration, and mice protected from systemic metabolic disease. The innate training phenotype of infection can be replicated by<span> </span></span><em>in vivo</em><span><span> </span>injection of SEA, and by exposure of bone marrow to SEA in culture prior to macrophage differentiation (Day 0). This protective phenotype is linked to increased chromatin accessibility of lipid and mitochondrial pathways in BMDM including Nrf1 and Tfam, as well as mitochondrial biogenesis. This work provides evidence that<span> </span></span><em>S. mansoni</em><span><span> </span>antigens induce a unique form of innate training inhibited by ovarian-derived hormones in females.</span></p>',
'date' => '2025-01-18',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2025.01.14.632838v1',
'doi' => 'https://doi.org/10.1101/2025.01.14.632838',
'modified' => '2025-01-31 10:55:42',
'created' => '2025-01-31 10:55:42',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '5117',
'name' => 'Dynamic PRC1–CBX8 stabilizes a porous structure of chromatin condensates',
'authors' => 'Uckelmann, M., et al.',
'description' => '<p><strong>Abstract</strong></p>
<div class="c-article-section__content" id="Abs1-content">
<p style="text-align: justify;">The compaction of chromatin is a prevalent paradigm in gene repression. Chromatin compaction is commonly thought to repress transcription by restricting chromatin accessibility. However, the spatial organization and dynamics of chromatin compacted by gene-repressing factors are unknown. Here, using cryo-electron tomography, we solved the three-dimensional structure of chromatin condensed by the polycomb repressive complex 1 (PRC1) in a complex with CBX8. PRC1-condensed chromatin is porous and stabilized through multivalent dynamic interactions of PRC1 with chromatin. Mechanistically, positively charged residues on the internally disordered regions of CBX8 mask negative charges on the DNA to stabilize the condensed state of chromatin. Within condensates, PRC1 remains dynamic while maintaining a static chromatin structure. In differentiated mouse embryonic stem cells, CBX8-bound chromatin remains accessible. These findings challenge the idea of rigidly compacted polycomb domains and instead provide a mechanistic framework for dynamic and accessible PRC1–chromatin condensates.</p>
</div>',
'date' => '2025-01-15',
'pmid' => 'https://www.nature.com/articles/s41594-024-01457-6',
'doi' => 'https://doi.org/10.1038/s41594-024-01457-6',
'modified' => '2025-04-25 13:20:40',
'created' => '2025-04-25 11:48:09',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 8 => array(
'id' => '5076',
'name' => 'Assessing the impact of whole genome duplication on gene expression and regulation during arachnid development',
'authors' => 'Madeleine E. Aase-Remedios et al.',
'description' => '<p id="p-2">Whole genome duplication (WGD) generates new genetic material that can contribute to the evolution of the regulation of developmental processes and phenotypic diversification. A WGD occurred in an ancestor of arachnopulmonates (spiders, scorpions, and their relatives), which provides an important independent comparison to WGDs in other animal lineages like those in vertebrates and horseshoe crabs. After WGD, arachnopulmonates retained many duplicated copies (ohnologues) of developmental genes including clusters of homeobox genes, many of which have been inferred to have undergone subfunctionalisation. However, there has been little systematic analysis of gene regulatory sequences and comparison of the expression of ohnologues versus their single-copy orthologues between arachnids. Here we compare the regions of accessible chromatin and gene expression of ohnologues and single-copy genes during three embryonic stages between an arachnopulmonate arachnid, the spider<span> </span><em>Parasteatoda tepidariorum</em>, and a non-arachnopulmonate arachnid, the harvestman<span> </span><em>Phalangium opilio</em>. We found that the expression of each spider ohnologue was lower than their single-copy orthologues in the harvestman providing evidence for subfunctionalisation. However, this was not reflected in a reduction in peaks of accessible chromatin because both spider ohnologues and single-copy genes had more peaks than the harvestman genes. We also found peaks of open chromatin that increased in the late stage associated with activation of genes expressed later during embryogenesis in both species. Taken together, our study provides the first genome-wide comparison of gene regulatory sequences and gene expression in arachnids and thus provides new insights into the impact of the arachnopulmonate WGD.</p>
<div id="sec-1" class="subsection">
<p id="p-3"><strong>Significance statement</strong><span> </span>The comparison of independent WGD events is essential to understanding their evolutionary outcomes. Here we examine gene expression and chromatin accessibility during the development of two arachnids, one of which underwent an ancient WGD. Our data provide the first direct comparison of the regulation of ohnologues and single-copy genes in arachnids.</p>
</div>',
'date' => '2024-12-20',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2024.12.18.628675v1.full',
'doi' => 'https://doi.org/10.1101/2024.12.18.628675',
'modified' => '2025-02-27 17:24:20',
'created' => '2025-02-27 17:24:20',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 9 => array(
'id' => '5012',
'name' => 'Spatial organizations of heterochromatin underpin nuclear structural integrity of ventricular cardiomyocytes against mechanical stress',
'authors' => 'Keita Fujiwara et al.',
'description' => '<section id="author-highlights-abstract" property="abstract" typeof="Text" role="doc-abstract">
<h2 property="name">Highlights</h2>
<div id="abspara0020" role="paragraph">
<div id="ulist0010" role="list">
<div id="u0010" role="listitem">
<div class="content">
<div id="p0010" role="paragraph">Cardiomyocytes acquire characteristic spatial organizations of heterochromatin (SOH)</div>
</div>
</div>
<div id="u0015" role="listitem">
<div class="content">
<div id="p0015" role="paragraph">High levels of H2B-mCherry disrupt SOH, leading to nuclear elongation in cardiomyocytes</div>
</div>
</div>
<div id="u0020" role="listitem">
<div class="content">
<div id="p0020" role="paragraph">SOH disruption minimally impacts gene expression despite loosening global genome structure</div>
</div>
</div>
<div id="u0025" role="listitem">
<div class="content">
<div id="p0025" role="paragraph">SOH alter with aging, leading to nuclear deformation in ventricular cardiomyocytes</div>
</div>
</div>
</div>
</div>
</section>
<section id="author-abstract" property="abstract" typeof="Text" role="doc-abstract">
<h2 property="name">Summary</h2>
<div id="abspara0010" role="paragraph">Cardiomyocyte (CM) nuclei are constantly exposed to mechanical stress, but how they maintain their nuclear shape remains unknown. In this study, we found that ventricular CM nuclei acquire characteristic prominent spatial organizations of heterochromatin (SOH), which are disrupted by high-level expression of H2B-mCherry in mice. SOH disruption was associated with nuclear softening, leading to extreme elongation and rupture under unidirectional mechanical stress. Loosened chromatin then leaks into the cytosol, causing severe inflammation and cardiac dysfunction. Although SOH disruption was accompanied by loosened higher-order genomic structures, the change in gene expression before nuclear deformation was mild, suggesting that SOH play major roles in nuclear structural integrity. Aged CM nuclei consistently exhibited scattered SOH and marked elongation. Furthermore, we provide mechanistic insight into the development and maintenance of SOH driven by chromatin compaction and condensate formation. These results highlight SOH as a safeguard of nuclear shape and genomic integrity against mechanical stress.</div>
</section>',
'date' => '2024-12-09',
'pmid' => 'https://www.cell.com/cell-reports/fulltext/S2211-1247(24)01399-8',
'doi' => '10.1016/j.celrep.2024.115048',
'modified' => '2024-12-12 15:04:47',
'created' => '2024-12-12 15:04:47',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 10 => array(
'id' => '4984',
'name' => 'DNA demethylation triggers cell free DNA release in colorectal cancer cells',
'authors' => 'Valeria Pessei et al.',
'description' => '<h3 class="c-article__sub-heading" data-test="abstract-sub-heading">Background</h3>
<p>Liquid biopsy based on cell-free DNA (cfDNA) analysis holds significant promise as a minimally invasive approach for the diagnosis, genotyping, and monitoring of solid malignancies. Human tumors release cfDNA in the bloodstream through a combination of events, including cell death, active and passive release. However, the precise mechanisms leading to cfDNA shedding remain to be characterized. Addressing this question in patients is confounded by several factors, such as tumor burden extent, anatomical and vasculature barriers, and release of nucleic acids from normal cells. In this work, we exploited cancer models to dissect basic mechanisms of DNA release.</p>
<h3 class="c-article__sub-heading" data-test="abstract-sub-heading">Methods</h3>
<p>We measured cell loss ratio, doubling time, and cfDNA release in the supernatant of a colorectal cancer (CRC) cell line collection (<i>N</i> = 76) representative of the molecular subtypes previously identified in cancer patients. Association analyses between quantitative parameters of cfDNA release, cell proliferation, and molecular features were evaluated. Functional experiments were performed to test the impact of modulating DNA methylation on cfDNA release.</p>
<h3 class="c-article__sub-heading" data-test="abstract-sub-heading">Results</h3>
<p>Higher levels of supernatant cfDNA were significantly associated with slower cell cycling and increased cell death. In addition, a higher cfDNA shedding was found in non-CpG Island Methylator Phenotype (CIMP) models. These results indicate a positive correlation between lower methylation and increased cfDNA levels. To explore this further, we exploited methylation microarrays to identify a subset of probes significantly associated with cfDNA shedding and derive a methylation signature capable of discriminating high from low cfDNA releasers. We applied this signature to an independent set of 176 CRC cell lines and patient derived organoids to select 14 models predicted to be low or high releasers. The methylation profile successfully predicted the amount of cfDNA released in the supernatant. At the functional level, genetic ablation of DNA methyl-transferases increased chromatin accessibility and DNA fragmentation, leading to increased cfDNA release in isogenic CRC cell lines. Furthermore, in vitro treatment of five low releaser CRC cells with a demethylating agent was able to induce a significant increase in cfDNA shedding.</p>
<h3 class="c-article__sub-heading" data-test="abstract-sub-heading">Conclusions</h3>
<p>Methylation status of cancer cell lines contributes to the variability of cfDNA shedding in vitro. Changes in methylation pattern are associated with cfDNA release levels and might be exploited to increase sensitivity of liquid biopsy assays.</p>',
'date' => '2024-10-09',
'pmid' => 'https://link.springer.com/article/10.1186/s13073-024-01386-5',
'doi' => 'https://doi.org/10.1186/s13073-024-01386-5',
'modified' => '2024-10-14 08:56:24',
'created' => '2024-10-14 08:56:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 11 => array(
'id' => '4967',
'name' => 'Temporal and spatial niche partitioning in a retrotransposon community of the Drosophila genome',
'authors' => 'Varoqui M. et al.',
'description' => '<p><span>Transposable elements (TEs), widespread genetic parasites, pose potential threats to the stability of their host genomes. Hence, the interactions observed today between TEs and their host genomes, as well as among the different TE species coexisting in the same host, likely reflect those that did not lead to the extinction of either the host or the TEs. It is not clear to what extent the expression and integration steps of the TE replication cycles are involved in this ‘peaceful’ coexistence. Here, we show that four Drosophila LTR RetroTransposable Elements (LTR-RTEs), although sharing the same overall integration mechanism, preferentially integrate into distinct open chromatin domains of the host germline. Notably, the differential expressions of the gtwin and ZAM LTR-RTEs in ovarian and embryonic somatic tissues, respectively, result in differential integration timings and targeting of accessible chromatin landscapes that differ between early and late embryonic nuclei, highlighting connections between temporal and spatial LTR-RTEs niche partitionings.</span></p>',
'date' => '2024-08-16',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2024.08.14.607943v1',
'doi' => 'https://doi.org/10.1101/2024.08.14.607943',
'modified' => '2024-09-02 10:29:44',
'created' => '2024-09-02 10:29:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 12 => array(
'id' => '5093',
'name' => 'Galectin 3-binding protein (LGALS3BP) depletion attenuates hepatic fibrosis by reducing transforming growth factor-β1 (TGF-β1) availability and inhibits hepatocarcinogenesis',
'authors' => 'Dae-Hwan Kim et al.',
'description' => '<section class="article-section__content" id="cac212600-sec-0010">
<h3 class="article-section__sub-title section1" id="cac212600-sec-0010-title">Background</h3>
<p>Increased Galectin 3-binding protein (LGALS3BP) serum levels have been used to assess hepatic fibrosis stages and the severity of hepatocellular carcinoma (HCC). Considering the crucial role of transforming growth factor-β1 (TGF-β1) in the emergence of these diseases, the present study tested the hypothesis that LGALS3BP regulates the TGF-β1 signaling pathway.</p>
</section>
<section class="article-section__content" id="cac212600-sec-0020">
<h3 class="article-section__sub-title section1" id="cac212600-sec-0020-title">Methods</h3>
<p>The expression levels of<span> </span><i>LGALS3BP</i><span> </span>and<span> </span><i>TGFB1</i><span> </span>were analyzed in patients with metabolic dysfunction-associated steatohepatitis (MASH) and HCC. Multiple omics techniques, such as RNA-sequencing, transposase-accessible chromatin-sequencing assay, and liquid chromatography-tandem mass spectrometry proteomics, were used to identify the regulatory mechanisms for the LGALS3BP-TGF-β1 axis. The effects of altered TGF-β1 signaling by LGALS3BP were investigated in conditional<span> </span><i>LGALS3BP</i>-knockin and<span> </span><i>LGALS3BP</i>-knockout mice.</p>
</section>
<section class="article-section__content" id="cac212600-sec-0030">
<h3 class="article-section__sub-title section1" id="cac212600-sec-0030-title">Results</h3>
<p>In patients with MASH and HCC, the levels of<span> </span><i>LGALS3BP</i><span> </span>and<span> </span><i>TGFB1</i><span> </span>exhibited positive correlations. Stimulation of LGALS3BP by the inflammatory cytokine interferon α in HCC cells or ectopic overexpression of LGALS3BP in hepatocytes promoted the expression levels of TGFB1. Aggravated fibrosis was observed in the livers of hepatocyte-specific<span> </span><i>LGALS3BP</i>-knockin mice, with increased TGFB1 levels. LGALS3BP directly bound to and assembled integrin αV, an integral mediator required for releasing active TGF-β1 from extracellular latent complex with the rearranged F-actin cytoskeleton. The released TGF-β1 activated JunB transcription factor, which in turn promoted the TGF-β1 positive feedback loop.<span> </span><i>LGALS3BP</i><span> </span>deletion in the hepatocytes downregulated TGF-β1 signaling and CCl<sub>4</sub><span> </span>induced fibrosis. Moreover,<span> </span><i>LGALS3BP</i><span> </span>depletion hindered hepatocarcinogenesis by limiting the availability of fibrogenic TGF-β1.</p>
</section>
<section class="article-section__content" id="cac212600-sec-0040">
<h3 class="article-section__sub-title section1" id="cac212600-sec-0040-title">Conclusion</h3>
<p>LGALS3BP plays a crucial role in hepatic fibrosis and carcinogenesis by controlling the TGF-β1 signaling pathway, making it a promising therapeutic target in TGF-β1-related diseases.</p>
</section>',
'date' => '2024-07-28',
'pmid' => 'https://onlinelibrary.wiley.com/doi/full/10.1002/cac2.12600',
'doi' => ' https://doi.org/10.1002/cac2.12600',
'modified' => '2025-03-27 16:59:26',
'created' => '2025-03-27 16:59:26',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 13 => array(
'id' => '5091',
'name' => 'Patho-transcriptomic analysis of invasive mucinous adenocarcinoma of the lung (IMA): comparison with lung adenocarcinoma with signet ring cell features (SRCC)',
'authors' => 'William D. Stuart et al.',
'description' => '<div id="sec-1" class="subsection">
<p id="p-3"><strong>Background</strong><span> </span>Invasive mucinous adenocarcinoma (IMA) comprises ∼5% of lung adenocarcinoma. There is no effective therapy for IMA when surgical resection is not possible. IMA is sometimes confused with adenocarcinoma with signet ring cell features (SRCC) pathologically since both adenocarcinomas feature tumor cells with abundant intracellular mucin. The molecular mechanisms by which such mucin-producing lung adenocarcinomas develop remain unknown.</p>
</div>
<div id="sec-2" class="subsection">
<p id="p-4"><strong>Methods</strong><span> </span>Using a Visium spatial transcriptomics approach, we analyzed IMA and compared it with SRCC patho-transcriptomically. Combining spatial transcriptomics data with<span> </span><em>in vitro</em><span> </span>studies using RNA-seq and ChIP-seq, we assessed downstream targets of transcription factors HNF4A and SPDEF that are highly expressed in IMA and/or SRCC.</p>
</div>
<div id="sec-3" class="subsection">
<p id="p-5"><strong>Results</strong><span> </span>Spatial transcriptomics analysis indicated that there are 6 distinct cell clusters in IMA and SRCC. Notably, two clusters (C1 and C3) of mucinous tumor cells exist in both adenocarcinomas albeit at a different ratio. Importantly, a portion of genes (e.g.,<span> </span><em>NKX2-1</em>,<span> </span><em>GKN1</em>,<span> </span><em>HNF4A</em><span> </span>and<span> </span><em>FOXA3</em>) are distinctly expressed while some mucous-related genes (e.g.,<span> </span><em>SPDEF</em><span> </span>and<span> </span><em>FOXA2</em>) are expressed in both adenocarcinomas. We determined that HNF4A induces<span> </span><em>MUC3A/B</em><span> </span>and<span> </span><em>TM4SF4</em><span> </span>and that BI 6015, an HNF4A antagonist, suppressed the growth of IMA cells. Using mutant SPDEF that is associated with COVID-19, we also determined that an intact DNA-binding domain of SPDEF is required for SPDEF-mediated induction of mucin genes (<em>MUC5AC</em>,<span> </span><em>MUC5B</em><span> </span>and<span> </span><em>AGR2</em>). Additionally, we found that XMU-MP-1, a SPDEF inhibitor, suppressed the growth of IMA cells.</p>
</div>
<div id="sec-4" class="subsection">
<p id="p-6"><strong>Conclusion</strong><span> </span>These results revealed that IMA and SRCC contain heterogenous tumor cell types, some of which are targetable.</p>
</div>',
'date' => '2024-06-17',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2024.06.13.598839v1.full',
'doi' => 'https://doi.org/10.1101/2024.06.13.598839',
'modified' => '2025-03-27 16:08:19',
'created' => '2025-03-27 16:08:19',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 14 => array(
'id' => '5094',
'name' => 'Transient loss of Polycomb components induces an epigenetic cancer fate',
'authors' => 'V. Parreno et al.',
'description' => '<p><span>Although cancer initiation and progression are generally associated with the accumulation of somatic mutations</span><sup><a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 1" title="McGranahan, N. & Swanton, C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 27, 15–26 (2015)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR1" id="ref-link-section-d121858e719">1</a>,<a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 2" title="Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR2" id="ref-link-section-d121858e722">2</a></sup><span>, substantial epigenomic alterations underlie many aspects of tumorigenesis and cancer susceptibility</span><sup><a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" title="Brock, A., Chang, H. & Huang, S. Non-genetic heterogeneity—a mutation-independent driving force for the somatic evolution of tumours. Nat. Rev. Genet. 10, 336–342 (2009)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR3" id="ref-link-section-d121858e726">3</a>,<a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" title="Flavahan, W. A., Gaskell, E. & Bernstein, B. E. Epigenetic plasticity and the hallmarks of cancer. Science 357, eaal2380 (2017)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR4" id="ref-link-section-d121858e726_1">4</a>,<a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" title="Marine, J. C., Dawson, S. J. & Dawson, M. A. Non-genetic mechanisms of therapeutic resistance in cancer. Nat. Rev. Cancer 20, 743–756 (2020)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR5" id="ref-link-section-d121858e726_2">5</a>,<a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 6" title="Timp, W. & Feinberg, A. P. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat. Rev. Cancer 13, 497–510 (2013)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR6" id="ref-link-section-d121858e729">6</a></sup><span>, suggesting that genetic mechanisms might not be the only drivers of malignant transformation</span><sup><a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 7" title="Teixeira, V. H. et al. Deciphering the genomic, epigenomic, and transcriptomic landscapes of pre-invasive lung cancer lesions. Nat. Med. 25, 517–525 (2019)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR7" id="ref-link-section-d121858e733">7</a></sup><span>. However, whether purely non-genetic mechanisms are sufficient to initiate tumorigenesis irrespective of mutations has been unknown. Here, we show that a transient perturbation of transcriptional silencing mediated by Polycomb group proteins is sufficient to induce an irreversible switch to a cancer cell fate in<span> </span></span><i>Drosophila</i><span>. This is linked to the irreversible derepression of genes that can drive tumorigenesis, including members of the JAK–STAT signalling pathway and<span> </span></span><i>zfh1</i><span>, the fly homologue of the<span> </span></span><i>ZEB1</i><span><span> </span>oncogene, whose aberrant activation is required for Polycomb perturbation-induced tumorigenesis. These data show that a reversible depletion of Polycomb proteins can induce cancer in the absence of driver mutations, suggesting that tumours can emerge through epigenetic dysregulation leading to inheritance of altered cell fates.</span></p>',
'date' => '2024-04-24',
'pmid' => 'https://www.nature.com/articles/s41586-024-07328-w',
'doi' => 'https://doi.org/10.1038/s41586-024-07328-w',
'modified' => '2025-03-27 17:00:44',
'created' => '2025-03-27 17:00:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 15 => array(
'id' => '4933',
'name' => 'Systematic mapping of TF-mediated cell fate changes by a pooled induction coupled with scRNA-seq and multi-omics approaches',
'authors' => 'Lee M. et al.',
'description' => '<p><span>Transcriptional regulation controls cellular functions through interactions between transcription factors (TFs) and their chromosomal targets. However, understanding the fate conversion potential of multiple TFs in an inducible manner remains limited. Here, we introduce iTF-seq as a method for identifying individual TFs that can alter cell fate toward specific lineages at a single-cell level. iTF-seq enables time course monitoring of transcriptome changes, and with biotinylated individual TFs, it provides a multi-omics approach to understanding the mechanisms behind TF-mediated cell fate changes. Our iTF-seq study in mouse embryonic stem cells identified multiple TFs that trigger rapid transcriptome changes indicative of differentiation within a day of induction. Moreover, cells expressing these potent TFs often show a slower cell cycle and increased cell death. Further analysis using bioChIP-seq revealed that GCM1 and OTX2 act as pioneer factors and activators by increasing gene accessibility and activating the expression of lineage specification genes during cell fate conversion. iTF-seq has utility in both mapping cell fate conversion and understanding cell fate conversion mechanisms.</span></p>',
'date' => '2024-04-05',
'pmid' => 'https://pubmed.ncbi.nlm.nih.gov/38580401/',
'doi' => '10.1101/gr.277926.123',
'modified' => '2024-04-09 00:19:18',
'created' => '2024-04-09 00:19:18',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 16 => array(
'id' => '4895',
'name' => 'Protocol to isolate nuclei from Chlamydomonas reinhardtii for ATAC sequencing',
'authors' => 'Santhanagopalan I. et al.',
'description' => '<p class="section-title u-h4 u-margin-l-top u-margin-xs-bottom">Highlights</p>
<div id="abssec0020">
<ul class="list">
<li class="react-xocs-list-item">
<p id="p0010">Optimized isolation of nuclei from the green model alga<span> </span><em>Chlamydomonas reinhardtii</em></p>
</li>
<li class="react-xocs-list-item">
<p id="p0010"><em></em></p>
<p id="p0015">Tag-free isolation from both cell-walled and cell wall-deficient algae strains</p>
</li>
<li class="react-xocs-list-item">
<p id="p0015"></p>
<p id="p0020">Key steps for an effective and fast isolation and quantification procedure of nuclei</p>
</li>
<li class="react-xocs-list-item">
<p><span class="list-label"></span>Extracts at a quality suitable for ATAC-sequencing</p>
</li>
</ul>
</div>',
'date' => '2024-03-15',
'pmid' => 'https://www.sciencedirect.com/science/article/pii/S2666166723007311',
'doi' => 'https://doi.org/10.1016/j.xpro.2023.102764',
'modified' => '2024-02-09 12:37:32',
'created' => '2024-01-22 13:39:58',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 17 => array(
'id' => '4917',
'name' => 'Interplay between coding and non-coding regulation drives the Arabidopsis seed-to-seedling transition',
'authors' => 'Trembley B.J.M. et al.',
'description' => '<p><span>Translation of seed stored mRNAs is essential to trigger germination. However, when RNAPII re-engages RNA synthesis during the seed-to-seedling transition has remained in question. Combining csRNA-seq, ATAC-seq and smFISH in </span><i>Arabidopsis thaliana</i><span><span> </span>we demonstrate that active transcription initiation is detectable during the entire germination process. Features of non-coding regulation such as dynamic changes in chromatin accessible regions, antisense transcription, as well as bidirectional non-coding promoters are widespread throughout the Arabidopsis genome. We show that sensitivity to exogenous ABSCISIC ACID (ABA) during germination depends on proximal promoter accessibility at ABA-responsive genes. Moreover, we provide genetic validation of the existence of divergent transcription in plants. Our results reveal that active enhancer elements are transcribed producing non-coding enhancer RNAs (eRNAs) as widely documented in metazoans. In sum, this study defining the extent and role of coding and non-coding transcription during key stages of germination expands our understanding of transcriptional mechanisms underlying plant developmental transitions.</span></p>',
'date' => '2024-02-26',
'pmid' => 'https://www.nature.com/articles/s41467-024-46082-5',
'doi' => 'https://doi.org/10.1038/s41467-024-46082-5',
'modified' => '2024-02-29 11:56:56',
'created' => '2024-02-29 11:56:56',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 18 => array(
'id' => '4910',
'name' => 'The transcriptional regulatory network modulating human trophoblast stem cells to extravillous trophoblast differentiation',
'authors' => 'Kim M. et al.',
'description' => '<p><span>During human pregnancy, extravillous trophoblasts play crucial roles in placental invasion into the maternal decidua and spiral artery remodeling. However, regulatory factors and their action mechanisms modulating human extravillous trophoblast specification have been unknown. By analyzing dynamic changes in transcriptome and enhancer profile during human trophoblast stem cell to extravillous trophoblast differentiation, we define stage-specific regulators, including an early-stage transcription factor, TFAP2C, and multiple late-stage transcription factors. Loss-of-function studies confirm the requirement of all transcription factors identified for adequate differentiation, and we reveal that the dynamic changes in the levels of TFAP2C are essential. Notably, TFAP2C pre-occupies the regulatory elements of the inactive extravillous trophoblast-active genes during the early stage of differentiation, and the late-stage transcription factors directly activate extravillous trophoblast-active genes, including themselves as differentiation further progresses, suggesting sequential actions of transcription factors assuring differentiation. Our results reveal stage-specific transcription factors and their inter-connected regulatory mechanisms modulating extravillous trophoblast differentiation, providing a framework for understanding early human placentation and placenta-related complications.</span></p>',
'date' => '2024-02-12',
'pmid' => 'https://www.nature.com/articles/s41467-024-45669-2',
'doi' => 'https://doi.org/10.1038/s41467-024-45669-2',
'modified' => '2024-02-15 10:43:52',
'created' => '2024-02-15 10:43:52',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 19 => array(
'id' => '4928',
'name' => 'DMSO derives Trophectoderm and Clonal Blastoid from Single Human Pluripotent Stem Cell',
'authors' => 'Alsolami S. et al.',
'description' => '<p><span>Human naïve pluripotent stem cells (nPSCs) can differentiate into extra-embryonic trophectoderm (TE), a critical step in the generation of the integrated embryo model termed blastoid. The current paradigm of blastoid generation necessitates the aggregation of dozens of nPSCs treated with multiple small molecule inhibitors, growth factors, or genetic modifications to initiate TE differentiation. The presence of complex crosstalk among pathways and cellular heterogeneity in these models complicates mechanistic study and genetic screens. Here, we show that a single small molecule, dimethyl sulfoxide (DMSO), potently induces TE differentiation in basal medium without pharmacological and genetic perturbations. DMSO enhances blastoid generation and, more importantly, is sufficient for blastoid generation by itself. DMSO blastoids resemble blastocysts in morphology and lineage composition. DMSO induces blastoid formation through PKC signaling and cell cycle regulation. Lastly, DMSO enables single nPSC-derived clonal blastoids, which could facilitate genetic screens for mechanistic understanding of human embryogenesis.</span></p>',
'date' => '2024-01-01',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2023.12.31.573770v1.abstract',
'doi' => 'https://doi.org/10.1101/2023.12.31.573770',
'modified' => '2024-03-27 15:18:04',
'created' => '2024-03-27 15:18:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 20 => array(
'id' => '4887',
'name' => 'In vitro production of cat-restricted Toxoplasma pre-sexual stages',
'authors' => 'Antunes, A.V. et al.',
'description' => '<p><span>Sexual reproduction of </span><i>Toxoplasma gondii</i><span>, confined to the felid gut, remains largely uncharted owing to ethical concerns regarding the use of cats as model organisms. Chromatin modifiers dictate the developmental fate of the parasite during its multistage life cycle, but their targeting to stage-specific cistromes is poorly described</span><sup><a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 1" title="Farhat, D. C. et al. A MORC-driven transcriptional switch controls Toxoplasma developmental trajectories and sexual commitment. Nat. Microbiol. 5, 570–583 (2020)." href="https://www.nature.com/articles/s41586-023-06821-y#ref-CR1" id="ref-link-section-d277698175e527">1</a>,<a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 2" title="Bougdour, A. et al. Drug inhibition of HDAC3 and epigenetic control of differentiation in Apicomplexa parasites. J. Exp. Med. 206, 953–966 (2009)." href="https://www.nature.com/articles/s41586-023-06821-y#ref-CR2" id="ref-link-section-d277698175e530">2</a></sup><span>. Here we found that the transcription factors AP2XII-1 and AP2XI-2 operate during the tachyzoite stage, a hallmark of acute toxoplasmosis, to silence genes necessary for merozoites, a developmental stage critical for subsequent sexual commitment and transmission to the next host, including humans. Their conditional and simultaneous depletion leads to a marked change in the transcriptional program, promoting a full transition from tachyzoites to merozoites. These in vitro-cultured pre-gametes have unique protein markers and undergo typical asexual endopolygenic division cycles. In tachyzoites, AP2XII-1 and AP2XI-2 bind DNA as heterodimers at merozoite promoters and recruit MORC and HDAC3 (ref. </span><sup><a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 1" title="Farhat, D. C. et al. A MORC-driven transcriptional switch controls Toxoplasma developmental trajectories and sexual commitment. Nat. Microbiol. 5, 570–583 (2020)." href="https://www.nature.com/articles/s41586-023-06821-y#ref-CR1" id="ref-link-section-d277698175e534">1</a></sup><span>), thereby limiting chromatin accessibility and transcription. Consequently, the commitment to merogony stems from a profound epigenetic rewiring orchestrated by AP2XII-1 and AP2XI-2. Successful production of merozoites in vitro paves the way for future studies on<span> </span></span><i>Toxoplasma</i><span><span> </span>sexual development without the need for cat infections and holds promise for the development of therapies to prevent parasite transmission.</span></p>',
'date' => '2023-12-13',
'pmid' => 'https://www.nature.com/articles/s41586-023-06821-y',
'doi' => 'https://doi.org/10.1038/s41586-023-06821-y',
'modified' => '2023-12-18 10:40:50',
'created' => '2023-12-18 10:40:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 21 => array(
'id' => '4939',
'name' => 'Vaginal microbes alter epithelial transcriptomic and epigenomic modifications providing insight into the molecular mechanisms for susceptibility to adverse reproductive outcomes',
'authors' => 'Elovitz M. et al.',
'description' => '<p><span>The cervicovaginal microbiome is highly associated with women's health with microbial communities dominated by </span><i>Lactobacillus</i><span><span> </span>spp. being considered optimal. Conversely, a lack of lactobacilli and a high abundance of strict and facultative anaerobes including<span> </span></span><i>Gardnerella vaginalis</i><span>, have been associated with adverse reproductive outcomes. However, the molecular pathways modulated by microbe interactions with the cervicovaginal epithelia remain unclear. Using RNA-sequencing, we characterize the<span> </span></span><i>in vitro</i><span><span> </span>cervicovaginal epithelial transcriptional response to different vaginal bacteria and their culture supernatants. We showed that<span> </span></span><i>G. vaginalis</i><span><span> </span>upregulated genes were associated with an activated innate immune response including anti-microbial peptides and inflammasome pathways, represented by NLRP3-mediated increases in caspase-1, IL-1β and cell death. Cervicovaginal epithelial cells exposed to<span> </span></span><i>L. crispatus</i><span><span> </span>showed limited transcriptomic changes, while exposure to<span> </span></span><i>L. crispatus</i><span><span> </span>culture supernatants resulted in a shift in the epigenomic landscape of cervical epithelial cells. ATAC-sequencing confirmed epigenetic changes with reduced chromatin accessibility. This study reveals new insight into host-microbe interactions in the lower reproductive tract and suggest potential therapeutic strategies leveraging the vaginal microbiome to improve reproductive health.</span></p>',
'date' => '2023-11-16',
'pmid' => 'https://pubmed.ncbi.nlm.nih.gov/38014044/',
'doi' => '10.21203/rs.3.rs-3580132/v1',
'modified' => '2024-06-07 15:47:48',
'created' => '2024-06-07 15:47:48',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 22 => array(
'id' => '4927',
'name' => 'Inflammatory stress-mediated chromatin changes underlie dysfunction in endothelial cells',
'authors' => 'Liu H. et al.',
'description' => '<p><span>Inflammatory stresses underlie endothelial dysfunction and contribute to the development of chronic cardiovascular disorders such as atherosclerosis and vascular fibrosis. The initial transcriptional response of endothelial cells to pro-inflammatory cytokines such as TNF-alpha is well established. However, very few studies uncover the effects of inflammatory stresses on chromatin architecture. We used integrative analysis of ATAC-seq and RNA-seq data to investigate chromatin alterations in human endothelial cells in response to TNF-alpha and febrile-range heat stress exposure. Multi-omics data analysis suggests a correlation between the transcription of stress-related genes and endothelial dysfunction drivers with chromatin regions exhibiting differential accessibility. Moreover, microscopy identified the dynamics in the nuclear organization, specifically, the changes in a subset of heterochromatic nucleoli-associated chromatin domains, the centromeres. Upon inflammatory stress exposure, the centromeres decreased association with nucleoli in a p38-dependent manner and increased the number of transcripts from pericentromeric regions. Overall, we provide two lines of evidence that suggest chromatin alterations in vascular endothelial cells during inflammatory stresses.</span></p>',
'date' => '2023-10-16',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10614786/',
'doi' => '10.1101/2023.10.11.561959',
'modified' => '2024-03-27 15:15:21',
'created' => '2024-03-27 15:15:21',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 23 => array(
'id' => '4880',
'name' => 'Dynamic PRC1-CBX8 stabilizes a porous structure of chromatin condensates',
'authors' => 'Uckelmann M. et al.',
'description' => '<p><span>The compaction of chromatin is a prevalent paradigm in gene repression. Chromatin compaction is commonly thought to repress transcription by restricting chromatin accessibility. However, the spatial organisation and dynamics of chromatin compacted by gene-repressing factors are unknown. Using cryo-electron tomography, we solved the three dimensional structure of chromatin condensed by the Polycomb Repressive Complex 1 (PRC1) in a complex with CBX8. PRC1-condensed chromatin is porous and stabilised through multivalent dynamic interactions of PRC1 with chromatin. Within condensates, PRC1 remains dynamic while maintaining a static chromatin structure. In differentiated mouse embryonic stem cells, CBX8-bound chromatin remains accessible. These findings challenge the idea of rigidly compacted polycomb domains and instead provides a mechanistic framework for dynamic and accessible PRC1-chromatin condensates.</span></p>',
'date' => '2023-05-09',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2023.05.08.539931v1.abstract',
'doi' => 'https://doi.org/10.1101/2023.05.08.539931',
'modified' => '2023-11-10 15:43:37',
'created' => '2023-11-10 15:43:37',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 24 => array(
'id' => '4810',
'name' => 'UMSBP2 is chromatin remodeler that functions in regulation of geneexpression and suppression of antigenic variation in trypanosomes.',
'authors' => 'Soni A. et al.',
'description' => '<p><span>Universal Minicircle Sequence binding proteins (UMSBPs) are CCHC-type zinc-finger proteins that bind the single-stranded G-rich UMS sequence, conserved at the replication origins of minicircles in the kinetoplast DNA, the mitochondrial genome of kinetoplastids. Trypanosoma brucei UMSBP2 has been recently shown to colocalize with telomeres and to play an essential role in chromosome end protection. Here we report that TbUMSBP2 decondenses in vitro DNA molecules, which were condensed by core histones H2B, H4 or linker histone H1. DNA decondensation is mediated via protein-protein interactions between TbUMSBP2 and these histones, independently of its previously described DNA binding activity. Silencing of the TbUMSBP2 gene resulted in a significant decrease in the disassembly of nucleosomes in T. brucei chromatin, a phenotype that could be reverted, by supplementing the knockdown cells with TbUMSBP2. Transcriptome analysis revealed that silencing of TbUMSBP2 affects the expression of multiple genes in T. brucei, with a most significant effect on the upregulation of the subtelomeric variant surface glycoproteins (VSG) genes, which mediate the antigenic variation in African trypanosomes. These observations suggest that UMSBP2 is a chromatin remodeling protein that functions in the regulation of gene expression and plays a role in the control of antigenic variation in T. brucei.</span></p>',
'date' => '2023-05-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/37207337',
'doi' => '10.1093/nar/gkad402',
'modified' => '2023-06-15 08:54:17',
'created' => '2023-06-13 21:11:31',
'ProductsPublication' => array(
[maximum depth reached]
)
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
'id' => '2440',
'name' => 'ATAC-seq kit SDS US en',
'language' => 'en',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-US-en-1_0.pdf',
'countries' => 'US',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '2438',
'name' => 'ATAC-seq kit SDS GB en',
'language' => 'en',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-GB-en-1_0.pdf',
'countries' => 'GB',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '2437',
'name' => 'ATAC-seq kit SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-FR-fr-1_0.pdf',
'countries' => 'FR',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '2433',
'name' => 'ATAC-seq kit SDS BE fr',
'language' => 'fr',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-BE-fr-1_0.pdf',
'countries' => 'BE',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '2436',
'name' => 'ATAC-seq kit SDS ES es',
'language' => 'es',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-ES-es-1_0.pdf',
'countries' => 'ES',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '2439',
'name' => 'ATAC-seq kit SDS JP ja',
'language' => 'ja',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-JP-ja-1_0.pdf',
'countries' => 'JP',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '2435',
'name' => 'ATAC-seq kit SDS DE de',
'language' => 'de',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-DE-de-1_0.pdf',
'countries' => 'DE',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '2434',
'name' => 'ATAC-seq kit SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-BE-nl-1_0.pdf',
'countries' => 'BE',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
)
)
)
$meta_canonical = 'https://dev.diagenode.com/en/p/atac-seq-kit-24rxns'
$fromEmail = 'dev.diagenode.com'
$country = 'US'
$countries_allowed = array(
(int) 0 => 'CA',
(int) 1 => 'US',
(int) 2 => 'IE',
(int) 3 => 'GB',
(int) 4 => 'DK',
(int) 5 => 'NO',
(int) 6 => 'SE',
(int) 7 => 'FI',
(int) 8 => 'NL',
(int) 9 => 'BE',
(int) 10 => 'LU',
(int) 11 => 'FR',
(int) 12 => 'DE',
(int) 13 => 'CH',
(int) 14 => 'AT',
(int) 15 => 'ES',
(int) 16 => 'IT',
(int) 17 => 'PT'
)
$outsource = false
$other_formats = array(
(int) 0 => array(
'id' => '3193',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<p><a href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p><strong>ATAC-seq</strong>, Assay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the <strong>transposase Tn5</strong> which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
<p>The Diagenode’s <b>ATAC-</b><b>seq</b><b> kit </b>is based on a highly validated protocol optimized for <b>50,000 </b><b>cells</b><b> per </b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The primer indexes for multiplexing are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell </b><b>requirement</b><b>: </b><b>50,000 </b><b>cells / </b><b>rxn</b></li>
<li><b>Robust protocol </b>with <b>high reproducibility </b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong> and <b>efficient DNA capture </b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids </b><b>over-amplification </b></li>
<li>Allows adaptation/flexibility for <b>more challenging samples </b>to succeed with library prep.</li>
<li>Gives <strong>early indication</strong> if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p><span>Looking for ATAC-seq on tissue? Please, go to: </span><a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack: <a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"> 0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"> (Tn5 transposase) </a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"> Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '8 rxns',
'catalog_number' => 'C01080001',
'old_catalog_number' => '',
'sf_code' => 'C01080001-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '615',
'price_USD' => '615',
'price_GBP' => '535',
'price_JPY' => '110700',
'price_CNY' => '',
'price_AUD' => '1538',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-8rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment (C01080001)| Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2023-05-05 07:57:08',
'created' => '2021-06-24 16:47:47',
'ProductsGroup' => array(
'id' => '397',
'product_id' => '3193',
'group_id' => '349'
)
)
)
$pro = array(
'id' => '3193',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<p><a href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p><strong>ATAC-seq</strong>, Assay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the <strong>transposase Tn5</strong> which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
<p>The Diagenode’s <b>ATAC-</b><b>seq</b><b> kit </b>is based on a highly validated protocol optimized for <b>50,000 </b><b>cells</b><b> per </b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The primer indexes for multiplexing are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell </b><b>requirement</b><b>: </b><b>50,000 </b><b>cells / </b><b>rxn</b></li>
<li><b>Robust protocol </b>with <b>high reproducibility </b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong> and <b>efficient DNA capture </b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids </b><b>over-amplification </b></li>
<li>Allows adaptation/flexibility for <b>more challenging samples </b>to succeed with library prep.</li>
<li>Gives <strong>early indication</strong> if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p><span>Looking for ATAC-seq on tissue? Please, go to: </span><a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack: <a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"> 0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"> (Tn5 transposase) </a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"> Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '8 rxns',
'catalog_number' => 'C01080001',
'old_catalog_number' => '',
'sf_code' => 'C01080001-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '615',
'price_USD' => '615',
'price_GBP' => '535',
'price_JPY' => '110700',
'price_CNY' => '',
'price_AUD' => '1538',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-8rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment (C01080001)| Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2023-05-05 07:57:08',
'created' => '2021-06-24 16:47:47',
'ProductsGroup' => array(
'id' => '397',
'product_id' => '3193',
'group_id' => '349'
)
)
$edit = ''
$testimonials = ''
$featured_testimonials = ''
$related_products = ''
$rrbs_service = array(
(int) 0 => (int) 1894,
(int) 1 => (int) 1895
)
$chipseq_service = array(
(int) 0 => (int) 2683,
(int) 1 => (int) 1835,
(int) 2 => (int) 1836,
(int) 3 => (int) 2684,
(int) 4 => (int) 1838,
(int) 5 => (int) 1839,
(int) 6 => (int) 1856
)
$labelize = object(Closure) {
}
$old_catalog_number = ''
$country_code = 'US'
$other_format = array(
'id' => '3193',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<p><a href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p><strong>ATAC-seq</strong>, Assay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the <strong>transposase Tn5</strong> which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
<p>The Diagenode’s <b>ATAC-</b><b>seq</b><b> kit </b>is based on a highly validated protocol optimized for <b>50,000 </b><b>cells</b><b> per </b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The primer indexes for multiplexing are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell </b><b>requirement</b><b>: </b><b>50,000 </b><b>cells / </b><b>rxn</b></li>
<li><b>Robust protocol </b>with <b>high reproducibility </b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong> and <b>efficient DNA capture </b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids </b><b>over-amplification </b></li>
<li>Allows adaptation/flexibility for <b>more challenging samples </b>to succeed with library prep.</li>
<li>Gives <strong>early indication</strong> if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p><span>Looking for ATAC-seq on tissue? Please, go to: </span><a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack: <a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"> 0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"> (Tn5 transposase) </a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"> Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '8 rxns',
'catalog_number' => 'C01080001',
'old_catalog_number' => '',
'sf_code' => 'C01080001-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '615',
'price_USD' => '615',
'price_GBP' => '535',
'price_JPY' => '110700',
'price_CNY' => '',
'price_AUD' => '1538',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-8rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment (C01080001)| Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2023-05-05 07:57:08',
'created' => '2021-06-24 16:47:47',
'ProductsGroup' => array(
'id' => '397',
'product_id' => '3193',
'group_id' => '349'
)
)
$label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>'
$document = array(
'id' => '1139',
'name' => 'ATAC-seq Kit - Manual',
'description' => '<p class="p1">Gene expression is carefully regulated in the cells in order to manage a wide range of biological functions. The structure of chromatin is quite dynamic and contributes to this crucial regulatory process.</p>
<p class="p1">ATAC-seq, Assay for Transposase-Accessible Chromatin, followed by nextgeneration sequencing, is a key technology to easily identify the “open” regions of the chromatin, which are usually associated with permissive gene expression. Indeed, the nuclei of the samples are incubated with a transposase, and only the genomic regions associated with open chromatin will be accessible to this transposase. During the process those regions will be cut and sequencing adaptors will be added, allowing their sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only., giving a map of the chromatin status in the whole genome of the sample.</p>
<p class="p1">The Diagenode’s ATAC-seq kit is based on a highly validated protocol, used for years in our Epigenomics Profiling Services offer and takes advantage of many successful Diagenode’s tools, such as the loaded Tagmentase (Tn5 transposase), the MicroChIP DiaPure Columns and the Primer indexes for tagmented libraries kits.</p>',
'image_id' => null,
'type' => 'Manual',
'url' => 'files/products/kits/atacseq-kit-manual.pdf',
'slug' => 'atacseq-kit-manual',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-06-25 09:51:59',
'created' => '2021-06-25 09:51:59',
'ProductsDocument' => array(
'id' => '3130',
'product_id' => '3194',
'document_id' => '1139'
)
)
$sds = array(
'id' => '2434',
'name' => 'ATAC-seq kit SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-BE-nl-1_0.pdf',
'countries' => 'BE',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
'id' => '4140',
'product_id' => '3194',
'safety_sheet_id' => '2434'
)
)
$publication = array(
'id' => '4810',
'name' => 'UMSBP2 is chromatin remodeler that functions in regulation of geneexpression and suppression of antigenic variation in trypanosomes.',
'authors' => 'Soni A. et al.',
'description' => '<p><span>Universal Minicircle Sequence binding proteins (UMSBPs) are CCHC-type zinc-finger proteins that bind the single-stranded G-rich UMS sequence, conserved at the replication origins of minicircles in the kinetoplast DNA, the mitochondrial genome of kinetoplastids. Trypanosoma brucei UMSBP2 has been recently shown to colocalize with telomeres and to play an essential role in chromosome end protection. Here we report that TbUMSBP2 decondenses in vitro DNA molecules, which were condensed by core histones H2B, H4 or linker histone H1. DNA decondensation is mediated via protein-protein interactions between TbUMSBP2 and these histones, independently of its previously described DNA binding activity. Silencing of the TbUMSBP2 gene resulted in a significant decrease in the disassembly of nucleosomes in T. brucei chromatin, a phenotype that could be reverted, by supplementing the knockdown cells with TbUMSBP2. Transcriptome analysis revealed that silencing of TbUMSBP2 affects the expression of multiple genes in T. brucei, with a most significant effect on the upregulation of the subtelomeric variant surface glycoproteins (VSG) genes, which mediate the antigenic variation in African trypanosomes. These observations suggest that UMSBP2 is a chromatin remodeling protein that functions in the regulation of gene expression and plays a role in the control of antigenic variation in T. brucei.</span></p>',
'date' => '2023-05-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/37207337',
'doi' => '10.1093/nar/gkad402',
'modified' => '2023-06-15 08:54:17',
'created' => '2023-06-13 21:11:31',
'ProductsPublication' => array(
'id' => '6971',
'product_id' => '3194',
'publication_id' => '4810'
)
)
$externalLink = ' <a href="https://www.ncbi.nlm.nih.gov/pubmed/37207337" target="_blank"><i class="fa fa-external-link"></i></a>'CustomErrorHandler::handleError() - APP/Lib/CustomErrorHandler.php, line 9
include - APP/View/Products/view.ctp, line 755
View::_evaluate() - CORE/Cake/View/View.php, line 971
View::_render() - CORE/Cake/View/View.php, line 933
View::render() - CORE/Cake/View/View.php, line 473
Controller::render() - CORE/Cake/Controller/Controller.php, line 963
ProductsController::slug() - APP/Controller/ProductsController.php, line 1055
ReflectionMethod::invokeArgs() - [internal], line ??
Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491
Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193
Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167
[main] - APP/webroot/index.php, line 118
(Notice): Undefined variable: message [APP/View/Products/view.ctp, line 755]Code Context $errorType = self::getErrorType($code);
self::logToDatabase( $errorType, $description, $file, $line);
return parent::handleError( $errorType, $description, $file, $line, $context);
$viewFile = '/var/www/dev.diagenode.com/app/View/Products/view.ctp'
$dataForView = array(
'language' => 'en',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'meta_title' => 'ATAC-seq kit for open chromatin assessment C01080001 | Diagenode ',
'product' => array(
'Product' => array(
'id' => '3194',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<div class="row">
<div class="small-12 medium-8 large-8 columns">
<p><a class="tiny details button" href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf">Download the Manual</a></p>
<p><strong>ATAC-seq</strong>, Assay for<span> </span><strong>T</strong>ransposase-<strong>A</strong>ccessible<span> </span><strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the<span> </span><strong>transposase Tn5</strong><span> </span>which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
</div>
<div class="small-12 medium-4 large-4 columns"><center>
<script>// <![CDATA[
var date = new Date(); var heure = date.getHours(); var jour = date.getDay(); var semaine = Math.floor(date.getDate() / 7) + 1; if (jour === 2 && ( (heure >= 9 && heure < 9.5) || (heure >= 18 && heure < 18.5) )) { document.write('<a href="https://us02web.zoom.us/j/85467619762"><img src="https://www.diagenode.com/img/epicafe-ON.gif"></a>'); } else { document.write('<a href="https://go.diagenode.com/l/928883/2023-04-26/3kq1v"><img src="https://www.diagenode.com/img/epicafe-OFF.png"></a>'); }
// ]]></script>
</center></div>
</div>
<p>Diagenode’s<span> </span><b>ATAC-</b><b>seq</b><b><span> </span>kit<span> </span></b>is based on a highly validated protocol optimized for<span> </span><b>50,000<span> </span></b><b>cells</b><b><span> </span>per<span> </span></b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The <a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">primer indexes for multiplexing</a> are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell<span> </span></b><b>requirement</b><b>:<span> </span></b><b>50,000<span> </span></b><b>cells /<span> </span></b><b>rxn</b></li>
<li><b>Robust protocol<span> </span></b>with<span> </span><b>high reproducibility<span> </span></b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong><span> </span>and<span> </span><b>efficient DNA capture<span> </span></b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids<span> </span></b><b>over-amplification</b></li>
<li>Allows adaptation/flexibility for<span> </span><b>more challenging samples<span> </span></b>to succeed with library prep.</li>
<li>Gives<span> </span><strong>early indication</strong><span> </span>if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p>Looking for ATAC-seq on tissue? Please, go to: <a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack:<span> </span><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"><span> </span>0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"><span> </span>(Tn5 transposase)<span> </span></a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"><span> </span>Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '24 rxns',
'catalog_number' => 'C01080002',
'old_catalog_number' => '',
'sf_code' => 'C01080002-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '1740',
'price_USD' => '1745',
'price_GBP' => '1485',
'price_JPY' => '313200',
'price_CNY' => '',
'price_AUD' => '4362',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-24rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment C01080001 | Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2025-04-17 10:29:58',
'created' => '2021-07-01 15:46:58',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => null,
'name' => null,
'description' => null,
'clonality' => null,
'isotype' => null,
'lot' => null,
'concentration' => null,
'reactivity' => null,
'type' => null,
'purity' => null,
'classification' => null,
'application_table' => null,
'storage_conditions' => null,
'storage_buffer' => null,
'precautions' => null,
'uniprot_acc' => null,
'slug' => null,
'meta_keywords' => null,
'meta_description' => null,
'modified' => null,
'created' => null,
'select_label' => null
),
'Slave' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Group' => array(
'Group' => array(
[maximum depth reached]
),
'Master' => array(
[maximum depth reached]
),
'Product' => array(
[maximum depth reached]
)
),
'Related' => array(),
'Application' => array(),
'Category' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Document' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
),
(int) 8 => array(
[maximum depth reached]
),
(int) 9 => array(
[maximum depth reached]
),
(int) 10 => array(
[maximum depth reached]
),
(int) 11 => array(
[maximum depth reached]
),
(int) 12 => array(
[maximum depth reached]
),
(int) 13 => array(
[maximum depth reached]
),
(int) 14 => array(
[maximum depth reached]
),
(int) 15 => array(
[maximum depth reached]
),
(int) 16 => array(
[maximum depth reached]
),
(int) 17 => array(
[maximum depth reached]
),
(int) 18 => array(
[maximum depth reached]
),
(int) 19 => array(
[maximum depth reached]
),
(int) 20 => array(
[maximum depth reached]
),
(int) 21 => array(
[maximum depth reached]
),
(int) 22 => array(
[maximum depth reached]
),
(int) 23 => array(
[maximum depth reached]
),
(int) 24 => array(
[maximum depth reached]
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
)
)
),
'meta_canonical' => 'https://dev.diagenode.com/en/p/atac-seq-kit-24rxns',
'fromEmail' => 'dev.diagenode.com'
)
$language = 'en'
$meta_keywords = ''
$meta_description = 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility'
$meta_title = 'ATAC-seq kit for open chromatin assessment C01080001 | Diagenode '
$product = array(
'Product' => array(
'id' => '3194',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<div class="row">
<div class="small-12 medium-8 large-8 columns">
<p><a class="tiny details button" href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf">Download the Manual</a></p>
<p><strong>ATAC-seq</strong>, Assay for<span> </span><strong>T</strong>ransposase-<strong>A</strong>ccessible<span> </span><strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the<span> </span><strong>transposase Tn5</strong><span> </span>which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
</div>
<div class="small-12 medium-4 large-4 columns"><center>
<script>// <![CDATA[
var date = new Date(); var heure = date.getHours(); var jour = date.getDay(); var semaine = Math.floor(date.getDate() / 7) + 1; if (jour === 2 && ( (heure >= 9 && heure < 9.5) || (heure >= 18 && heure < 18.5) )) { document.write('<a href="https://us02web.zoom.us/j/85467619762"><img src="https://www.diagenode.com/img/epicafe-ON.gif"></a>'); } else { document.write('<a href="https://go.diagenode.com/l/928883/2023-04-26/3kq1v"><img src="https://www.diagenode.com/img/epicafe-OFF.png"></a>'); }
// ]]></script>
</center></div>
</div>
<p>Diagenode’s<span> </span><b>ATAC-</b><b>seq</b><b><span> </span>kit<span> </span></b>is based on a highly validated protocol optimized for<span> </span><b>50,000<span> </span></b><b>cells</b><b><span> </span>per<span> </span></b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The <a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">primer indexes for multiplexing</a> are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell<span> </span></b><b>requirement</b><b>:<span> </span></b><b>50,000<span> </span></b><b>cells /<span> </span></b><b>rxn</b></li>
<li><b>Robust protocol<span> </span></b>with<span> </span><b>high reproducibility<span> </span></b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong><span> </span>and<span> </span><b>efficient DNA capture<span> </span></b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids<span> </span></b><b>over-amplification</b></li>
<li>Allows adaptation/flexibility for<span> </span><b>more challenging samples<span> </span></b>to succeed with library prep.</li>
<li>Gives<span> </span><strong>early indication</strong><span> </span>if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p>Looking for ATAC-seq on tissue? Please, go to: <a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack:<span> </span><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"><span> </span>0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"><span> </span>(Tn5 transposase)<span> </span></a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"><span> </span>Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '24 rxns',
'catalog_number' => 'C01080002',
'old_catalog_number' => '',
'sf_code' => 'C01080002-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '1740',
'price_USD' => '1745',
'price_GBP' => '1485',
'price_JPY' => '313200',
'price_CNY' => '',
'price_AUD' => '4362',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-24rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment C01080001 | Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2025-04-17 10:29:58',
'created' => '2021-07-01 15:46:58',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => null,
'name' => null,
'description' => null,
'clonality' => null,
'isotype' => null,
'lot' => null,
'concentration' => null,
'reactivity' => null,
'type' => null,
'purity' => null,
'classification' => null,
'application_table' => null,
'storage_conditions' => null,
'storage_buffer' => null,
'precautions' => null,
'uniprot_acc' => null,
'slug' => null,
'meta_keywords' => null,
'meta_description' => null,
'modified' => null,
'created' => null,
'select_label' => null
),
'Slave' => array(
(int) 0 => array(
'id' => '349',
'name' => 'C01080002',
'product_id' => '3194',
'modified' => '2021-07-01 15:47:50',
'created' => '2021-07-01 15:47:50'
)
),
'Group' => array(
'Group' => array(
'id' => '349',
'name' => 'C01080002',
'product_id' => '3194',
'modified' => '2021-07-01 15:47:50',
'created' => '2021-07-01 15:47:50'
),
'Master' => array(
'id' => '3194',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<div class="row">
<div class="small-12 medium-8 large-8 columns">
<p><a class="tiny details button" href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf">Download the Manual</a></p>
<p><strong>ATAC-seq</strong>, Assay for<span> </span><strong>T</strong>ransposase-<strong>A</strong>ccessible<span> </span><strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the<span> </span><strong>transposase Tn5</strong><span> </span>which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
</div>
<div class="small-12 medium-4 large-4 columns"><center>
<script>// <![CDATA[
var date = new Date(); var heure = date.getHours(); var jour = date.getDay(); var semaine = Math.floor(date.getDate() / 7) + 1; if (jour === 2 && ( (heure >= 9 && heure < 9.5) || (heure >= 18 && heure < 18.5) )) { document.write('<a href="https://us02web.zoom.us/j/85467619762"><img src="https://www.diagenode.com/img/epicafe-ON.gif"></a>'); } else { document.write('<a href="https://go.diagenode.com/l/928883/2023-04-26/3kq1v"><img src="https://www.diagenode.com/img/epicafe-OFF.png"></a>'); }
// ]]></script>
</center></div>
</div>
<p>Diagenode’s<span> </span><b>ATAC-</b><b>seq</b><b><span> </span>kit<span> </span></b>is based on a highly validated protocol optimized for<span> </span><b>50,000<span> </span></b><b>cells</b><b><span> </span>per<span> </span></b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The <a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">primer indexes for multiplexing</a> are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell<span> </span></b><b>requirement</b><b>:<span> </span></b><b>50,000<span> </span></b><b>cells /<span> </span></b><b>rxn</b></li>
<li><b>Robust protocol<span> </span></b>with<span> </span><b>high reproducibility<span> </span></b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong><span> </span>and<span> </span><b>efficient DNA capture<span> </span></b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids<span> </span></b><b>over-amplification</b></li>
<li>Allows adaptation/flexibility for<span> </span><b>more challenging samples<span> </span></b>to succeed with library prep.</li>
<li>Gives<span> </span><strong>early indication</strong><span> </span>if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p>Looking for ATAC-seq on tissue? Please, go to: <a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack:<span> </span><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"><span> </span>0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"><span> </span>(Tn5 transposase)<span> </span></a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"><span> </span>Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '24 rxns',
'catalog_number' => 'C01080002',
'old_catalog_number' => '',
'sf_code' => 'C01080002-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '1740',
'price_USD' => '1745',
'price_GBP' => '1485',
'price_JPY' => '313200',
'price_CNY' => '',
'price_AUD' => '4362',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-24rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment C01080001 | Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2025-04-17 10:29:58',
'created' => '2021-07-01 15:46:58'
),
'Product' => array(
(int) 0 => array(
[maximum depth reached]
)
)
),
'Related' => array(),
'Application' => array(),
'Category' => array(
(int) 0 => array(
'id' => '13',
'position' => '2',
'parent_id' => null,
'name' => 'Chromatin studies',
'description' => '<div class="row">
<div class="small-12 medium-8 large-8 columns"><br />
<p>Chromatin structure plays a key role in regulating gene expression by allowing DNA accessibility to transcriptional machinery and transcription factors. The packaging of DNA into nucleosomes forms a closed structure that is not highly accessible to transcriptional elements whereas the open nucleosome structure allows DNA to be accessible. Diagenode offers a number of solutions to help you analyze chromatin and the role of transcriptional machinery including ChIP kits, ChIPmentation kits, antibodies, pA-Tn5 and ATAC-seq kits.</p>
</div>
<div class="small-12 medium-4 large-4 columns"><center>
<script>// <![CDATA[
var date = new Date(); var heure = date.getHours(); var jour = date.getDay(); var semaine = Math.floor(date.getDate() / 7) + 1; if (jour === 2 && ( (heure >= 9 && heure < 9.5) || (heure >= 18 && heure < 18.5) )) { document.write('<a href="https://us02web.zoom.us/j/85467619762"><img src="https://www.diagenode.com/img/epicafe-ON.gif"></a>'); } else { document.write('<a href="https://go.diagenode.com/l/928883/2023-04-26/3kq1v"><img src="https://www.diagenode.com/img/epicafe-OFF.png"></a>'); }
// ]]></script>
</center></div>
</div>
<p><a href="https://www.diagenode.com/en/p/uchipmentation-for-histones-24-rxns"><img src="https://www.diagenode.com/img/banners/b-microchip-category.png" /></a></p>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<div id="portal" class="main-portal">
<div class="portal-inner"><nav class="portal-nav">
<ul class="tips-menu">
<li><a href="#workflow" class="tips portal button" style="background: #13b29c; color: #f3fbfa;">Chromatin immunoprecipitation</a></li>
<li><a href="https://www.diagenode.com/en/categories/chromatin-ip-chipmentation" class="tips portal button">ChIPmentation</a></li>
<li><a href="https://www.diagenode.com/en/categories/antibodies
" class="tips portal button">Antibodies</a></li>
<li><a href="https://www.diagenode.com/en/categories/cutandtag" class="tips portal button">pA-Tn5</a></li>
<li><a href="https://www.diagenode.com/en/categories/atac-seq" class="tips portal button">ATAC-seq</a></li>
<li><a href="https://www.diagenode.com/en/categories/cutandtag" class="tips portal button">CUT&Tag</a></li>
</ul>
</nav></div>
</div>
<p>Chromatin immunoprecipitation (ChIP) determines the location of DNA binding sites on the genome for a protein of interest, giving insights into gene expression regulation. ChIP involves the selective enrichment of a chromatin fraction containing a specific antigen. Antibodies that recognize a protein or protein modification are used to determine the relative abundance of that antigen at a specific locus or loci. ChIP can be used to compare enrichment of proteins, map protein modifications, or quantify a protein modification during a time course.</p>
<span class="anchor" id="workflow"></span>
<h4><span style="font-weight: 400;">The ChIP workflow</span></h4>
<ul class="accordion" data-accordion="">
<li class="accordion-navigation"><img src="https://www.diagenode.com/img/categories/kits_chromatin_function/website-chip-workflow.jpg" />
<div id="chip_workflow" class="content">
<div class="row">
<table>
<tbody>
<tr>
<td width="50%">
<h3 class="text-center">Step by step workflow</h3>
</td>
<td width="50%">
<h3 class="text-center">Optimal solution from Diagenode</h3>
</td>
</tr>
<tr>
<td colspan="2"></td>
</tr>
<tr valign="top">
<td>
<p class="lead text-center"><strong>1.</strong><span> </span>Crosslink to bind proteins to DNA</p>
<center><img src="https://www.diagenode.com/img/chip/step-by-step-1-workflow.png" width="192" height="24" /></center></td>
<td>
<ul class="arrow">
<li><a href="https://www.diagenode.com/p/chip-cross-link-gold-600-ul">ChIP Cross-link Gold</a></li>
</ul>
</td>
</tr>
<tr>
<td colspan="2"></td>
</tr>
<tr valign="top">
<td>
<p class="lead text-center"><strong>2.</strong><span> </span>Shear DNA</p>
<center><img src="https://www.diagenode.com/img/chip/step-by-step-2-workflow.png" width="194" height="80" /></center></td>
<td>
<ul class="arrow">
<li><a href="https://www.diagenode.com/p/bioruptor-pico-sonication-device#">Bioruptor<sup>®</sup><span> </span>Pico Sonication device</a></li>
<li><a href="https://www.diagenode.com/categories/chromatin-shearing">Shearing optimization reagent</a></li>
</ul>
</td>
</tr>
<tr>
<td colspan="2"></td>
</tr>
<tr valign="top">
<td>
<p class="lead text-center"><strong>3.</strong><span> </span>Immunoprecipitate with specific antibody</p>
<center><img src="https://www.diagenode.com/img/chip/step-by-step-3-workflow.png" width="47" height="51" /></center></td>
<td>
<ul class="arrow">
<li><a href="https://www.diagenode.com/categories/chromatin-immunoprecipitation">ChIP-seq and ChIP-qPCR kits</a><span> </span>for transcription factors, histones, low inputs, plants</li>
<li><a href="https://www.diagenode.com/categories/chip-grade-antibodies">ChIP</a><span> </span>and<span> </span><a href="https://www.diagenode.com/categories/chip-seq-grade-antibodies">ChIP-seq grade</a><span> </span>antibodies</li>
<li><a href="https://www.diagenode.com/categories/ip-star">Automation available</a></li>
</ul>
</td>
</tr>
<tr>
<td colspan="2"></td>
</tr>
<tr valign="top">
<td>
<p class="lead text-center"><strong>4.</strong><span> </span>Reverse crosslinks and purify</p>
<center><img src="https://www.diagenode.com/img/chip/step-by-step-4-workflow.png" width="194" height="80" /></center></td>
<td>
<ul class="arrow">
<li>Diagenode's ChIP kits (contain optimal purification modules)</li>
</ul>
</td>
</tr>
<tr>
<td colspan="2"></td>
</tr>
<tr valign="top">
<td>
<p class="lead text-center"><strong>5.</strong><span> </span><a href="https://www.diagenode.com/en/p/ideal-chip-qpcr-kit">ChIP-qPCR</a><span> </span>or ChIP-seq library preparation</p>
</td>
<td>
<ul class="arrow">
<li>ChIP-seq:</li>
</ul>
<ul style="list-style-type: square;">
<li style="margin-left: 40px; font-size: 1.09rem;"><a href="https://www.diagenode.com/p/microplex-library-preparation-kit-v2-x48-12-indices-48-rxns">MicroPlex Library Preparation for 50 pg - 5 ng</a></li>
<li style="margin-left: 40px; font-size: 1.09rem;"><a href="https://www.diagenode.com/p/ideal-library-preparation-kit-x24-incl-index-primer-set-1-24-rxns">iDeal Library Preparation for > 5 ng</a></li>
</ul>
<ul class="arrow">
<li>ChIP qPCR</li>
</ul>
</td>
</tr>
</tbody>
</table>
</div>
</div>
</li>
</ul>
<p></p>
<h4><span style="font-weight: 400;">Products for chromatin study</span></h4>
<p><span style="font-weight: 400;"></span></p>
<p style="padding-left: 30px;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/crosslinking.png" alt="" width="35" height="35" /> Crosslinking<br /></b></span></strong><span style="font-weight: 400;">Efficient solution for protein-protein fixation.</span><strong><span style="font-weight: 400;"><b><span style="font-weight: 400;"> <a href="../p/chip-cross-link-gold-600-ul
">Read more</a></span></b><br /></span></strong></p>
<p style="padding-left: 30px;"><strong><img src="https://www.diagenode.com/img/applications/chromatin-shearing.png" alt="" width="35" height="35" /> Chromatin shearing<br /></strong><strong><span style="font-weight: 400;">Perfectly sheared chromatin is critical for ChIP success.<span> </span><br /></span></strong><strong><a href="../categories/chromatin-shearing"><span style="font-weight: 400;">Read more</span></a><span style="font-weight: 400;"><span> </span>about solutions for successful chromatin preparation.<span> </span><br /></span></strong><strong><span style="font-weight: 400;"><a href="../categories/bioruptor-shearing-device">Read more</a><span> </span>about<span> </span></span><span style="font-weight: 400;">Bioruptor<span> </span></span><span style="font-weight: 400;">sonication.</span></strong></p>
<p style="padding-left: 30px;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/immunoprecipitation.png" width="35" height="35" caption="false" /> Chromatin immunoprecipitation<br /></b></span></strong><span style="font-weight: 400;">Immunoprecipitation solutions for histone and transcription factor ChIP-qPCR and ChIP-seq for low inputs, plants, and animals including automated solutions</span><strong><span style="font-weight: 400;"><b><span style="font-weight: 400;">. <a href="../categories/chromatin-immunoprecipitation">Read more</a></span></b><br /></span></strong></p>
<p style="padding-left: 30px;"><strong><img src="https://www.diagenode.com/img/applications/ChIPmentation.png" alt="" width="35" height="35" /> ChIPmentation<br /><span style="font-weight: 400;">ChIPmentation, an exclusive technology, is an end-to-end ChIP-seq solution for low and difficult inputs. <a href="../categories/chromatin-ip-chipmentation">Read more</a></span><br /></strong></p>
<p style="padding-left: 30px;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/ChIP-antibodies.png" alt="" width="35" height="35" /> ChIP and ChIP-seq antibodies<br /></b></span></strong><span style="font-weight: 400;">ChIP-grade antibodies are essential for success. <a href="../categories/antibodies">Learn more</a></span><span style="font-weight: 400;"><br /></span></p>
<p style="padding-left: 30px;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/ChIP-qPCR.png" width="35" height="35" caption="false" /> Primer pairs<br /></b></span></strong><span style="font-weight: 400;">Highly specific primer pairs for the amplification of the specific genomic regions. <a href="../categories/primer-pairs">Read more</a></span><span style="font-weight: 400;"><br /></span></p>
<p style="padding-left: 30px;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/cutandtag.png" width="35" height="35" caption="false" /> CUT&Tag solutions<br /></b></span></strong><span style="font-weight: 400;">An alternative to ChIP-seq that combines antibody-targeted controlled cleavage by a protein A-Tn5 fusion with NGS to identify the binding sites of DNA-associated proteins. <a href="../categories/cutandtag">Read more</a></span><span style="font-weight: 400;"><br /></span></p>
<p style="padding-left: 30px;"><span style="font-weight: 400;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/dna-purification.png" width="35" height="35" caption="false" /> DNA purification<br /></b></span></strong><span style="font-weight: 400;"><a href="../categories/dna-and-rna-purification">Read more</a> about solutions for DNA purification</span></span></p>
<p style="padding-left: 30px;"><span style="font-weight: 400;"><span style="font-weight: 400;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/ChIP-seq.png" width="35" height="35" caption="false" /> Library preparation for ChIP-seq<br /></b></span></strong><span style="font-weight: 400;">Optimized solutions for the library preparation from low DNA input. <a href="../categories/library-preparation-for-ChIP-seq">Read more</a></span></span></span></p>
<p style="padding-left: 30px;"><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/ChIP-automation.png" alt="" width="35" height="35" /> ChIP and ChIP-seq automation<br /></b>Reproducibility, optimization simplicity, no variability. <a href="../categories/epigenetic-automation">Learn more</a></span></p>
<div class="row" style="margin-top: 32px;">
<div class="small-12 medium-10 large-9 small-centered columns">
<div class="radius panel" style="background-color: #fff;">
<p class="text-justify">We offer <a href="https://www.diagenode.com/en/categories/chromatin-immunoprecipitation" target="_blank">complete ChIP kits</a> or <strong>individual kit components</strong> from antibodies, buffers, beads, chromatin shearing, and purification reagents. With the ChIP Kit Customizer, you have complete flexibility on which components you want from our validated ChIP kits.</p>
<div class="center" style="text-align: center;"><a href="../pages/chip-kit-customizer-1"><img src="https://www.diagenode.com/img/banners/banner-customizer.png" alt="" /></a></div>
</div>
</div>
</div>
<h4>Chromatin resources</h4>
<h3>Posters</h3>
<ul>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/antibody-chipseq-qc-using-the-ipstar-compact-poster"><span style="font-weight: 400;">Understanding our antibody QC</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/antibodies-you-can-trust-poster"><span style="font-weight: 400;">High quality ChIP antibodies</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/chip-kit-results-with-true-microchip-kit-poster"><span style="font-weight: 400;">ChIP with only 10,000 cells</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/high-resolution-chipseq-profiles-with-ipstar-automated-platform-poster"><span style="font-weight: 400;">High resolution ChIP-seq using automation</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/bioinformatics-pipeline-for-chipseq-analyses"><span style="font-weight: 400;">ChIP-seq bioinformatics</span></a></li>
</ul>
<h3><span style="font-weight: 400;">Application notes</span></h3>
<ul>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/chipettor-application-note"><span style="font-weight: 400;">Simple semi-automaton for easy and inexpensive ChIP</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/chipseq-from-human-tumor-tissue"><span style="font-weight: 400;">Performing ChIP-seq on human tumor tissue</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/plant-chip-seq-application-note"><span style="font-weight: 400;">Plant ChIP-seq – a successful method</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/chip-seq-application-note"><span style="font-weight: 400;">Best workflow practices for low input ChIP</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/files/application_notes/AN-ChIP-Cas9-02_2018.pdf"><span style="font-weight: 400;">Optimize the selection of guide RNA by ChIP to keep CRISPR on-target</span></a></li>
</ul>
<h3>Publications related to ChIP</h3>
<ul>
<li><a href="https://www.diagenode.com/en/publications/view/3373">Corticosteroid receptors adopt distinct cyclical transcriptional signatures</a></li>
<li><a href="https://www.diagenode.com/en/publications/view/3347">Pro-inflammatory cytokine and high doses of ionizing radiation have similar effects on the expression of NF-kappaB-dependent genes</a></li>
<li><a href="https://www.diagenode.com/en/publications/view/3355">Functional dissection of Drosophila melanogaster SUUR protein influence on H3K27me3 profile</a></li>
</ul>
<h3><span style="font-weight: 400;">Brochures</span></h3>
<ul>
<li><a href="https://www.diagenode.com/files/brochures/Chromatin_Immunoprecipitation_Brochure.pdf"><b>Chromatin </b><span style="font-weight: 400;">products brochure</span></a></li>
<li><a href="https://www.diagenode.com/files/brochures/Epigenetic_Antibodies_Brochure.pdf"><span style="font-weight: 400;">Epigenetic<span> </span></span><b>Antibodies</b></a></li>
<li><a href="https://www.diagenode.com/files/brochures/Bioruptor_Sonicator_Brochure.pdf"><span style="font-weight: 400;">Bioruptor for<span> </span></span><b>chromatin shearing</b></a></li>
<li><a href="https://www.diagenode.com/files/brochures/IPStar_Automated_System_Brochure.pdf"><span style="font-weight: 400;">Automating<span> </span></span><b>ChIP</b><span style="font-weight: 400;"><span> </span>and<span> </span></span><b>ChIP-seq</b></a></li>
</ul>
</div>
</div>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => true,
'hide' => false,
'all_format' => false,
'is_antibody' => false,
'slug' => 'chromatin-function',
'cookies_tag_id' => null,
'meta_keywords' => 'Chromatin function,Chromatin shearing,Chromatin immunoprecipitation,Chromatin assembly',
'meta_description' => 'Diagenode offers a number of unique solutions and kits to make Diagenode offers chromatin immunoprecipitation kits have been designed to meet a wide variety of research needs and accessible',
'meta_title' => 'Chromatin Function Kit for Unique Solutions and Kits to make Chromatin Research| Diagenode',
'modified' => '2025-05-21 12:06:52',
'created' => '2015-05-27 05:22:15',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
)
),
'Document' => array(
(int) 0 => array(
'id' => '1139',
'name' => 'ATAC-seq Kit - Manual',
'description' => '<p class="p1">Gene expression is carefully regulated in the cells in order to manage a wide range of biological functions. The structure of chromatin is quite dynamic and contributes to this crucial regulatory process.</p>
<p class="p1">ATAC-seq, Assay for Transposase-Accessible Chromatin, followed by nextgeneration sequencing, is a key technology to easily identify the “open” regions of the chromatin, which are usually associated with permissive gene expression. Indeed, the nuclei of the samples are incubated with a transposase, and only the genomic regions associated with open chromatin will be accessible to this transposase. During the process those regions will be cut and sequencing adaptors will be added, allowing their sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only., giving a map of the chromatin status in the whole genome of the sample.</p>
<p class="p1">The Diagenode’s ATAC-seq kit is based on a highly validated protocol, used for years in our Epigenomics Profiling Services offer and takes advantage of many successful Diagenode’s tools, such as the loaded Tagmentase (Tn5 transposase), the MicroChIP DiaPure Columns and the Primer indexes for tagmented libraries kits.</p>',
'image_id' => null,
'type' => 'Manual',
'url' => 'files/products/kits/atacseq-kit-manual.pdf',
'slug' => 'atacseq-kit-manual',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-06-25 09:51:59',
'created' => '2021-06-25 09:51:59',
'ProductsDocument' => array(
[maximum depth reached]
)
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
'id' => '1819',
'name' => 'product/kits/atacseq-kit-picture.jpg',
'alt' => 'ATAC-seq kit',
'modified' => '2021-06-25 10:03:22',
'created' => '2021-06-25 10:03:22',
'ProductsImage' => array(
[maximum depth reached]
)
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
'id' => '5143',
'name' => 'CRAMP1 drives linker histone expression to enable Polycomb repression',
'authors' => 'Matthews, Rachael E et al.',
'description' => '<div class="abstract-content selected" id="eng-abstract">
<p style="text-align: justify;">In contrast to the well-understood role of core histones in DNA packaging, the function of the linker histone (H1) remains enigmatic. Challenging the prevailing view that linker histones are a general feature of heterochromatin, here we show a critical requirement for H1 in Polycomb repressive complex 2 (PRC2) function. A CRISPR-Cas9 genetic screen using a fluorescent PRC2 reporter identified an essential role for the poorly characterized gene CRAMP1 in PRC2-mediated repression. CRAMP1 localizes to the promoters of expressed H1 genes and positively regulates their transcription. CRAMP1 ablation simultaneously depletes all linker histones, which results in selective decompaction of H3K27me3-marked loci and derepression of PRC2 target genes without concomitant loss of PRC2 occupancy or enzymatic activity. Strikingly, we find that linker histones preferentially localize to genomic loci marked by H3K27me3 across diverse cell types and organisms. Altogether, these data demonstrate a prominent role for linker histones in epigenetic repression by PRC2.</p>
</div>',
'date' => '2025-06-10',
'pmid' => 'https://pubmed.ncbi.nlm.nih.gov/40516528/',
'doi' => '10.1016/j.molcel.2025.05.031',
'modified' => '2025-06-18 09:45:01',
'created' => '2025-06-18 09:45:01',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '5142',
'name' => 'Temporal and spatial niche partitioning in a retrotransposon community of the Drosophila melanogaster genome',
'authors' => 'Varoqui, Marion et al.',
'description' => '<div class="abstract" id="abstract">
<div class="abstract-content selected" id="eng-abstract">
<p style="text-align: justify;">Transposable elements (TEs) are genetic parasites that can potentially threaten the stability of the genomes they colonize. Nonetheless, TEs persist within genomes and are rarely fully eliminated, with diverse TE families coexisting in varing copy numbers. The TE replication strategies that enable host organisms to tolerate and accommodate the extensive diversity of TEs, while minimizing harm to the host and avoiding mutual competition among TEs, remain poorly understood. Here, by studying the spontaneous or experimental mobilization of four Drosophila LTR RetroTransposable Elements (LTR-RTEs), we reveal that each of them preferentially targets open chromatin regions characterized by specific epigenetic features. Among these, gtwin and ZAM are expressed in distinct cell types within female somatic gonadal tissues and inserted into the distinct accessible chromatin landscapes of the corresponding stages of embryogenesis. These findings suggest that individual LTR-RTEs exploit unique biological niches, enabling their coexistence within the tightly regulated ecosystem of the same host genome.</p>
</div>
</div>',
'date' => '2025-06-06',
'pmid' => 'https://pubmed.ncbi.nlm.nih.gov/40503684/',
'doi' => '10.1093/nar/gkaf516',
'modified' => '2025-06-18 09:43:08',
'created' => '2025-06-18 09:43:08',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '5128',
'name' => 'Cervicovaginal microbiome alters transcriptomic and epigenomic signatures across cervicovaginal epithelial barriers',
'authors' => 'Lauren Anton et al. ',
'description' => '<p><b>Background</b></p>
<p>The cervicovaginal microbiome plays a critical role in women's health, with microbial communities dominated by<span> </span><em>Lactobacillus</em><span> </span>species considered optimal. In contrast, the depletion of lactobacilli and the presence of a diverse array of strict and facultative anaerobes, such as<span> </span><em>Gardnerella vaginalis</em>, have been linked with adverse reproductive outcomes. Despite these associations, the molecular mechanisms by which host-microbial interactions modulate cervical and vaginal epithelial function remains poorly understood.</p>
<p><b>Results</b></p>
<p>In this study, we used RNA sequencing to characterize the transcriptional response of cervicovaginal epithelial cells exposed to the culture supernatants of common vaginal bacteria. Our findings revealed that<span> </span><em>G. vaginalis</em><span> </span>culture supernatants upregulate genes associated with an activated innate immune response and increased cell death. Conversely,<span> </span><em>Lactobacillus crispatus</em><span> </span>culture supernatants induced transcriptional changes indicative of epigenomic modeling in ectocervical epithelial cells. Epigenomic modification by<span> </span><em>L. crispatus</em>, was confirmed by ATAC-sequencing, which demonstrated reduced chromatin accessibility.</p>
<p><b>Conclusions</b></p>
<p>These results provide new insights into host-microbe interactions within the lower reproductive tract and suggests that modulating the vaginal microbiome could offer innovative therapeutic strategies to improve reproductive health.</p>',
'date' => '2025-05-07',
'pmid' => 'https://www.researchsquare.com/article/rs-6171614/v1',
'doi' => 'https://doi.org/10.21203/rs.3.rs-6171614/v1',
'modified' => '2025-05-12 10:30:24',
'created' => '2025-05-12 10:30:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '5125',
'name' => 'An ISWI-related chromatin remodeller regulates stage-specific gene expression in Toxoplasma gondii',
'authors' => 'Pachano, Belen et al.',
'description' => '<p style="text-align: justify;"><span>ATP-dependent chromatin remodellers are specialized multiprotein machines that organize the genome in eukaryotic cells and regulate its accessibility by repositioning, ejecting or modifying nucleosomes. However, their role in </span><i>Toxoplasma gondii</i><span><span> </span>is poorly understood. Here we show that<span> </span></span><i>T.</i><span> </span><i>gondii</i><span><span> </span>has evolved two divergent proteins within the imitation switch (ISWI) family:<span> </span></span><i>Tg</i><span>SNF2h and<span> </span></span><i>Tg</i><span>SNF2L.<span> </span></span><i>Tg</i><span>SNF2h specifically forms a core complex with the transcription factor AP2VIII-2 and the scaffold protein<span> </span></span><i>Tg</i><span>RFTS. Depletion of<span> </span></span><i>Tg</i><span>RFTS phenocopies the knockdown of<span> </span></span><i>Tg</i><span>SNF2h, restricting access to chromatin and altering local gene expression. At the genomic level,<span> </span></span><i>Tg</i><span>SNF2h insulates highly transcribed genes from silenced neighbours, ensuring stage-specific gene regulation. By modulating chromatin accessibility to transcription factors,<span> </span></span><i>Tg</i><span>SNF2h exerts epistatic control over MORC, a key regulator of sexual commitment. Our findings show that a specific ISWI complex orchestrates the partitioning of developmental genes and ensures transcriptional fidelity throughout the parasite life cycle.</span></p>',
'date' => '2025-04-11',
'pmid' => 'https://www.nature.com/articles/s41564-025-01980-2#citeas',
'doi' => 'https://doi.org/10.1038/s41564-025-01980-2',
'modified' => '2025-05-19 11:36:47',
'created' => '2025-05-06 15:49:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '5101',
'name' => 'Repression of oxidative phosphorylation by NR2F2, MTERF3 and GDF15 in human skin under high-glucose stress',
'authors' => 'S. Ley-Ngardigal et al.',
'description' => '<p><span>Lifestyle factors such as a Western diet or metabolic diseases like diabetes disrupt glucose homeostasis and induce stress responses, yet their impact on skin metabolism and structural integrity remains poorly understood. Here, we performed multiomic and bioenergetic analyses of human dermal fibroblasts (HDFs), human equivalent dermis (HED), human reconstructed skin (HRS), and skin explants from diabetic patients. We found that 12 mM glucose stress represses oxidative phosphorylation (OXPHOS) through a dual mechanism: the glucose-dependent nuclear receptor NR2F2 activates mitochondrial transcription termination factor 3 (MTERF3) while inhibiting growth-differentiation factor 15 (GDF15). Promoter assays revealed that MTERF3 is regulated by NR2F2 and MYCN, whereas GDF15 is modulated by NR2F2 and FOS. Consequently, OXPHOS proteins and mitochondrial respiration were suppressed, and MTERF3 overexpression additionally interfered with collagen biosynthesis. In contrast, GDF15 supplementation fully rescued hyperglycemia-induced bioenergetic and metabolomic alterations, suggesting a pharmacological strategy to mitigate hyperglycemic damage in the skin. Finally, silencing GDF15 or TFAM impaired fibroblast haptotaxis and skin reconstruction, underscoring the crucial role of mitochondrial energetics in dermal structure and function. Collectively, these findings identify the NR2F2–MTERF3–GDF15 axis as a key mediator of OXPHOS suppression and highlight a potential therapeutic target to preserve skin integrity under hyperglycemic stress.</span></p>',
'date' => '2025-03-27',
'pmid' => 'https://www.sciencedirect.com/science/article/pii/S2213231725001260',
'doi' => 'https://doi.org/10.1016/j.redox.2025.103613',
'modified' => '2025-03-31 13:44:10',
'created' => '2025-03-31 13:44:10',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '5092',
'name' => 'CRISPR screening identifies regulators of enhancer-mediated androgen receptor transcription in advanced prostate cancer',
'authors' => 'Rachel R. Xiang et al. ',
'description' => '<section id="author-highlights-abstract" property="abstract" typeof="Text" role="doc-abstract">
<h2 property="name"></h2>
</section>
<section id="author-abstract" property="abstract" typeof="Text" role="doc-abstract">
<div id="abspara0010" role="paragraph">Amplification of the androgen receptor (AR) locus is the most frequent alteration in metastatic castration-resistant prostate cancer (CRPC). Recently, it was discovered that an enhancer of the AR is co-amplified with the AR gene body and contributes to increased AR transcription and resistance to androgen deprivation therapy. However, the mechanism of enhancer activation in advanced disease is unknown. Here, we used CRISPR-Cas9 screening to identify transcription factors that bind to the AR enhancer and modulate enhancer-mediated AR transcription. We demonstrate that HOXB13, GATA2, and TFAP2C bind the AR enhancer in patient-derived xenografts and directly impact features associated with an active chromatin state. Interestingly, the AR enhancer belongs to a set of regulatory elements that require HOXB13 to maintain FOXA1 binding, further delineating the role of HOXB13 in CRPC. This work provides a framework to functionally identify<span> </span><i>trans</i>-acting factors required for the activation of disease-related noncoding regulatory elements.</div>
</section>',
'date' => '2025-02-14',
'pmid' => 'https://www.cell.com/cell-reports/fulltext/S2211-1247(25)00083-X',
'doi' => '10.1016/j.celrep.2025.115312',
'modified' => '2025-03-27 16:56:52',
'created' => '2025-03-27 16:54:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '5038',
'name' => 'Schistosoma mansoni antigen induced innate immune memory features mitochondrial biogenesis and can be inhibited by ovarian produced hormones',
'authors' => 'Juan Marcos Oviedo et al.',
'description' => '<p><span>We have previously identified that </span><em>S. mansoni</em><span><span> </span>infection induces a unique form of myeloid training that protects male but not female mice from high fat diet induced disease. Here we demonstrate that ovarian derived hormones account for this sex specific difference. Ovariectomy of females prior to infection permits metabolic reprogramming of the myeloid lineage, with BMDM exhibiting carbon source flexibility for cellular respiration, and mice protected from systemic metabolic disease. The innate training phenotype of infection can be replicated by<span> </span></span><em>in vivo</em><span><span> </span>injection of SEA, and by exposure of bone marrow to SEA in culture prior to macrophage differentiation (Day 0). This protective phenotype is linked to increased chromatin accessibility of lipid and mitochondrial pathways in BMDM including Nrf1 and Tfam, as well as mitochondrial biogenesis. This work provides evidence that<span> </span></span><em>S. mansoni</em><span><span> </span>antigens induce a unique form of innate training inhibited by ovarian-derived hormones in females.</span></p>',
'date' => '2025-01-18',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2025.01.14.632838v1',
'doi' => 'https://doi.org/10.1101/2025.01.14.632838',
'modified' => '2025-01-31 10:55:42',
'created' => '2025-01-31 10:55:42',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '5117',
'name' => 'Dynamic PRC1–CBX8 stabilizes a porous structure of chromatin condensates',
'authors' => 'Uckelmann, M., et al.',
'description' => '<p><strong>Abstract</strong></p>
<div class="c-article-section__content" id="Abs1-content">
<p style="text-align: justify;">The compaction of chromatin is a prevalent paradigm in gene repression. Chromatin compaction is commonly thought to repress transcription by restricting chromatin accessibility. However, the spatial organization and dynamics of chromatin compacted by gene-repressing factors are unknown. Here, using cryo-electron tomography, we solved the three-dimensional structure of chromatin condensed by the polycomb repressive complex 1 (PRC1) in a complex with CBX8. PRC1-condensed chromatin is porous and stabilized through multivalent dynamic interactions of PRC1 with chromatin. Mechanistically, positively charged residues on the internally disordered regions of CBX8 mask negative charges on the DNA to stabilize the condensed state of chromatin. Within condensates, PRC1 remains dynamic while maintaining a static chromatin structure. In differentiated mouse embryonic stem cells, CBX8-bound chromatin remains accessible. These findings challenge the idea of rigidly compacted polycomb domains and instead provide a mechanistic framework for dynamic and accessible PRC1–chromatin condensates.</p>
</div>',
'date' => '2025-01-15',
'pmid' => 'https://www.nature.com/articles/s41594-024-01457-6',
'doi' => 'https://doi.org/10.1038/s41594-024-01457-6',
'modified' => '2025-04-25 13:20:40',
'created' => '2025-04-25 11:48:09',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 8 => array(
'id' => '5076',
'name' => 'Assessing the impact of whole genome duplication on gene expression and regulation during arachnid development',
'authors' => 'Madeleine E. Aase-Remedios et al.',
'description' => '<p id="p-2">Whole genome duplication (WGD) generates new genetic material that can contribute to the evolution of the regulation of developmental processes and phenotypic diversification. A WGD occurred in an ancestor of arachnopulmonates (spiders, scorpions, and their relatives), which provides an important independent comparison to WGDs in other animal lineages like those in vertebrates and horseshoe crabs. After WGD, arachnopulmonates retained many duplicated copies (ohnologues) of developmental genes including clusters of homeobox genes, many of which have been inferred to have undergone subfunctionalisation. However, there has been little systematic analysis of gene regulatory sequences and comparison of the expression of ohnologues versus their single-copy orthologues between arachnids. Here we compare the regions of accessible chromatin and gene expression of ohnologues and single-copy genes during three embryonic stages between an arachnopulmonate arachnid, the spider<span> </span><em>Parasteatoda tepidariorum</em>, and a non-arachnopulmonate arachnid, the harvestman<span> </span><em>Phalangium opilio</em>. We found that the expression of each spider ohnologue was lower than their single-copy orthologues in the harvestman providing evidence for subfunctionalisation. However, this was not reflected in a reduction in peaks of accessible chromatin because both spider ohnologues and single-copy genes had more peaks than the harvestman genes. We also found peaks of open chromatin that increased in the late stage associated with activation of genes expressed later during embryogenesis in both species. Taken together, our study provides the first genome-wide comparison of gene regulatory sequences and gene expression in arachnids and thus provides new insights into the impact of the arachnopulmonate WGD.</p>
<div id="sec-1" class="subsection">
<p id="p-3"><strong>Significance statement</strong><span> </span>The comparison of independent WGD events is essential to understanding their evolutionary outcomes. Here we examine gene expression and chromatin accessibility during the development of two arachnids, one of which underwent an ancient WGD. Our data provide the first direct comparison of the regulation of ohnologues and single-copy genes in arachnids.</p>
</div>',
'date' => '2024-12-20',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2024.12.18.628675v1.full',
'doi' => 'https://doi.org/10.1101/2024.12.18.628675',
'modified' => '2025-02-27 17:24:20',
'created' => '2025-02-27 17:24:20',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 9 => array(
'id' => '5012',
'name' => 'Spatial organizations of heterochromatin underpin nuclear structural integrity of ventricular cardiomyocytes against mechanical stress',
'authors' => 'Keita Fujiwara et al.',
'description' => '<section id="author-highlights-abstract" property="abstract" typeof="Text" role="doc-abstract">
<h2 property="name">Highlights</h2>
<div id="abspara0020" role="paragraph">
<div id="ulist0010" role="list">
<div id="u0010" role="listitem">
<div class="content">
<div id="p0010" role="paragraph">Cardiomyocytes acquire characteristic spatial organizations of heterochromatin (SOH)</div>
</div>
</div>
<div id="u0015" role="listitem">
<div class="content">
<div id="p0015" role="paragraph">High levels of H2B-mCherry disrupt SOH, leading to nuclear elongation in cardiomyocytes</div>
</div>
</div>
<div id="u0020" role="listitem">
<div class="content">
<div id="p0020" role="paragraph">SOH disruption minimally impacts gene expression despite loosening global genome structure</div>
</div>
</div>
<div id="u0025" role="listitem">
<div class="content">
<div id="p0025" role="paragraph">SOH alter with aging, leading to nuclear deformation in ventricular cardiomyocytes</div>
</div>
</div>
</div>
</div>
</section>
<section id="author-abstract" property="abstract" typeof="Text" role="doc-abstract">
<h2 property="name">Summary</h2>
<div id="abspara0010" role="paragraph">Cardiomyocyte (CM) nuclei are constantly exposed to mechanical stress, but how they maintain their nuclear shape remains unknown. In this study, we found that ventricular CM nuclei acquire characteristic prominent spatial organizations of heterochromatin (SOH), which are disrupted by high-level expression of H2B-mCherry in mice. SOH disruption was associated with nuclear softening, leading to extreme elongation and rupture under unidirectional mechanical stress. Loosened chromatin then leaks into the cytosol, causing severe inflammation and cardiac dysfunction. Although SOH disruption was accompanied by loosened higher-order genomic structures, the change in gene expression before nuclear deformation was mild, suggesting that SOH play major roles in nuclear structural integrity. Aged CM nuclei consistently exhibited scattered SOH and marked elongation. Furthermore, we provide mechanistic insight into the development and maintenance of SOH driven by chromatin compaction and condensate formation. These results highlight SOH as a safeguard of nuclear shape and genomic integrity against mechanical stress.</div>
</section>',
'date' => '2024-12-09',
'pmid' => 'https://www.cell.com/cell-reports/fulltext/S2211-1247(24)01399-8',
'doi' => '10.1016/j.celrep.2024.115048',
'modified' => '2024-12-12 15:04:47',
'created' => '2024-12-12 15:04:47',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 10 => array(
'id' => '4984',
'name' => 'DNA demethylation triggers cell free DNA release in colorectal cancer cells',
'authors' => 'Valeria Pessei et al.',
'description' => '<h3 class="c-article__sub-heading" data-test="abstract-sub-heading">Background</h3>
<p>Liquid biopsy based on cell-free DNA (cfDNA) analysis holds significant promise as a minimally invasive approach for the diagnosis, genotyping, and monitoring of solid malignancies. Human tumors release cfDNA in the bloodstream through a combination of events, including cell death, active and passive release. However, the precise mechanisms leading to cfDNA shedding remain to be characterized. Addressing this question in patients is confounded by several factors, such as tumor burden extent, anatomical and vasculature barriers, and release of nucleic acids from normal cells. In this work, we exploited cancer models to dissect basic mechanisms of DNA release.</p>
<h3 class="c-article__sub-heading" data-test="abstract-sub-heading">Methods</h3>
<p>We measured cell loss ratio, doubling time, and cfDNA release in the supernatant of a colorectal cancer (CRC) cell line collection (<i>N</i> = 76) representative of the molecular subtypes previously identified in cancer patients. Association analyses between quantitative parameters of cfDNA release, cell proliferation, and molecular features were evaluated. Functional experiments were performed to test the impact of modulating DNA methylation on cfDNA release.</p>
<h3 class="c-article__sub-heading" data-test="abstract-sub-heading">Results</h3>
<p>Higher levels of supernatant cfDNA were significantly associated with slower cell cycling and increased cell death. In addition, a higher cfDNA shedding was found in non-CpG Island Methylator Phenotype (CIMP) models. These results indicate a positive correlation between lower methylation and increased cfDNA levels. To explore this further, we exploited methylation microarrays to identify a subset of probes significantly associated with cfDNA shedding and derive a methylation signature capable of discriminating high from low cfDNA releasers. We applied this signature to an independent set of 176 CRC cell lines and patient derived organoids to select 14 models predicted to be low or high releasers. The methylation profile successfully predicted the amount of cfDNA released in the supernatant. At the functional level, genetic ablation of DNA methyl-transferases increased chromatin accessibility and DNA fragmentation, leading to increased cfDNA release in isogenic CRC cell lines. Furthermore, in vitro treatment of five low releaser CRC cells with a demethylating agent was able to induce a significant increase in cfDNA shedding.</p>
<h3 class="c-article__sub-heading" data-test="abstract-sub-heading">Conclusions</h3>
<p>Methylation status of cancer cell lines contributes to the variability of cfDNA shedding in vitro. Changes in methylation pattern are associated with cfDNA release levels and might be exploited to increase sensitivity of liquid biopsy assays.</p>',
'date' => '2024-10-09',
'pmid' => 'https://link.springer.com/article/10.1186/s13073-024-01386-5',
'doi' => 'https://doi.org/10.1186/s13073-024-01386-5',
'modified' => '2024-10-14 08:56:24',
'created' => '2024-10-14 08:56:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 11 => array(
'id' => '4967',
'name' => 'Temporal and spatial niche partitioning in a retrotransposon community of the Drosophila genome',
'authors' => 'Varoqui M. et al.',
'description' => '<p><span>Transposable elements (TEs), widespread genetic parasites, pose potential threats to the stability of their host genomes. Hence, the interactions observed today between TEs and their host genomes, as well as among the different TE species coexisting in the same host, likely reflect those that did not lead to the extinction of either the host or the TEs. It is not clear to what extent the expression and integration steps of the TE replication cycles are involved in this ‘peaceful’ coexistence. Here, we show that four Drosophila LTR RetroTransposable Elements (LTR-RTEs), although sharing the same overall integration mechanism, preferentially integrate into distinct open chromatin domains of the host germline. Notably, the differential expressions of the gtwin and ZAM LTR-RTEs in ovarian and embryonic somatic tissues, respectively, result in differential integration timings and targeting of accessible chromatin landscapes that differ between early and late embryonic nuclei, highlighting connections between temporal and spatial LTR-RTEs niche partitionings.</span></p>',
'date' => '2024-08-16',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2024.08.14.607943v1',
'doi' => 'https://doi.org/10.1101/2024.08.14.607943',
'modified' => '2024-09-02 10:29:44',
'created' => '2024-09-02 10:29:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 12 => array(
'id' => '5093',
'name' => 'Galectin 3-binding protein (LGALS3BP) depletion attenuates hepatic fibrosis by reducing transforming growth factor-β1 (TGF-β1) availability and inhibits hepatocarcinogenesis',
'authors' => 'Dae-Hwan Kim et al.',
'description' => '<section class="article-section__content" id="cac212600-sec-0010">
<h3 class="article-section__sub-title section1" id="cac212600-sec-0010-title">Background</h3>
<p>Increased Galectin 3-binding protein (LGALS3BP) serum levels have been used to assess hepatic fibrosis stages and the severity of hepatocellular carcinoma (HCC). Considering the crucial role of transforming growth factor-β1 (TGF-β1) in the emergence of these diseases, the present study tested the hypothesis that LGALS3BP regulates the TGF-β1 signaling pathway.</p>
</section>
<section class="article-section__content" id="cac212600-sec-0020">
<h3 class="article-section__sub-title section1" id="cac212600-sec-0020-title">Methods</h3>
<p>The expression levels of<span> </span><i>LGALS3BP</i><span> </span>and<span> </span><i>TGFB1</i><span> </span>were analyzed in patients with metabolic dysfunction-associated steatohepatitis (MASH) and HCC. Multiple omics techniques, such as RNA-sequencing, transposase-accessible chromatin-sequencing assay, and liquid chromatography-tandem mass spectrometry proteomics, were used to identify the regulatory mechanisms for the LGALS3BP-TGF-β1 axis. The effects of altered TGF-β1 signaling by LGALS3BP were investigated in conditional<span> </span><i>LGALS3BP</i>-knockin and<span> </span><i>LGALS3BP</i>-knockout mice.</p>
</section>
<section class="article-section__content" id="cac212600-sec-0030">
<h3 class="article-section__sub-title section1" id="cac212600-sec-0030-title">Results</h3>
<p>In patients with MASH and HCC, the levels of<span> </span><i>LGALS3BP</i><span> </span>and<span> </span><i>TGFB1</i><span> </span>exhibited positive correlations. Stimulation of LGALS3BP by the inflammatory cytokine interferon α in HCC cells or ectopic overexpression of LGALS3BP in hepatocytes promoted the expression levels of TGFB1. Aggravated fibrosis was observed in the livers of hepatocyte-specific<span> </span><i>LGALS3BP</i>-knockin mice, with increased TGFB1 levels. LGALS3BP directly bound to and assembled integrin αV, an integral mediator required for releasing active TGF-β1 from extracellular latent complex with the rearranged F-actin cytoskeleton. The released TGF-β1 activated JunB transcription factor, which in turn promoted the TGF-β1 positive feedback loop.<span> </span><i>LGALS3BP</i><span> </span>deletion in the hepatocytes downregulated TGF-β1 signaling and CCl<sub>4</sub><span> </span>induced fibrosis. Moreover,<span> </span><i>LGALS3BP</i><span> </span>depletion hindered hepatocarcinogenesis by limiting the availability of fibrogenic TGF-β1.</p>
</section>
<section class="article-section__content" id="cac212600-sec-0040">
<h3 class="article-section__sub-title section1" id="cac212600-sec-0040-title">Conclusion</h3>
<p>LGALS3BP plays a crucial role in hepatic fibrosis and carcinogenesis by controlling the TGF-β1 signaling pathway, making it a promising therapeutic target in TGF-β1-related diseases.</p>
</section>',
'date' => '2024-07-28',
'pmid' => 'https://onlinelibrary.wiley.com/doi/full/10.1002/cac2.12600',
'doi' => ' https://doi.org/10.1002/cac2.12600',
'modified' => '2025-03-27 16:59:26',
'created' => '2025-03-27 16:59:26',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 13 => array(
'id' => '5091',
'name' => 'Patho-transcriptomic analysis of invasive mucinous adenocarcinoma of the lung (IMA): comparison with lung adenocarcinoma with signet ring cell features (SRCC)',
'authors' => 'William D. Stuart et al.',
'description' => '<div id="sec-1" class="subsection">
<p id="p-3"><strong>Background</strong><span> </span>Invasive mucinous adenocarcinoma (IMA) comprises ∼5% of lung adenocarcinoma. There is no effective therapy for IMA when surgical resection is not possible. IMA is sometimes confused with adenocarcinoma with signet ring cell features (SRCC) pathologically since both adenocarcinomas feature tumor cells with abundant intracellular mucin. The molecular mechanisms by which such mucin-producing lung adenocarcinomas develop remain unknown.</p>
</div>
<div id="sec-2" class="subsection">
<p id="p-4"><strong>Methods</strong><span> </span>Using a Visium spatial transcriptomics approach, we analyzed IMA and compared it with SRCC patho-transcriptomically. Combining spatial transcriptomics data with<span> </span><em>in vitro</em><span> </span>studies using RNA-seq and ChIP-seq, we assessed downstream targets of transcription factors HNF4A and SPDEF that are highly expressed in IMA and/or SRCC.</p>
</div>
<div id="sec-3" class="subsection">
<p id="p-5"><strong>Results</strong><span> </span>Spatial transcriptomics analysis indicated that there are 6 distinct cell clusters in IMA and SRCC. Notably, two clusters (C1 and C3) of mucinous tumor cells exist in both adenocarcinomas albeit at a different ratio. Importantly, a portion of genes (e.g.,<span> </span><em>NKX2-1</em>,<span> </span><em>GKN1</em>,<span> </span><em>HNF4A</em><span> </span>and<span> </span><em>FOXA3</em>) are distinctly expressed while some mucous-related genes (e.g.,<span> </span><em>SPDEF</em><span> </span>and<span> </span><em>FOXA2</em>) are expressed in both adenocarcinomas. We determined that HNF4A induces<span> </span><em>MUC3A/B</em><span> </span>and<span> </span><em>TM4SF4</em><span> </span>and that BI 6015, an HNF4A antagonist, suppressed the growth of IMA cells. Using mutant SPDEF that is associated with COVID-19, we also determined that an intact DNA-binding domain of SPDEF is required for SPDEF-mediated induction of mucin genes (<em>MUC5AC</em>,<span> </span><em>MUC5B</em><span> </span>and<span> </span><em>AGR2</em>). Additionally, we found that XMU-MP-1, a SPDEF inhibitor, suppressed the growth of IMA cells.</p>
</div>
<div id="sec-4" class="subsection">
<p id="p-6"><strong>Conclusion</strong><span> </span>These results revealed that IMA and SRCC contain heterogenous tumor cell types, some of which are targetable.</p>
</div>',
'date' => '2024-06-17',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2024.06.13.598839v1.full',
'doi' => 'https://doi.org/10.1101/2024.06.13.598839',
'modified' => '2025-03-27 16:08:19',
'created' => '2025-03-27 16:08:19',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 14 => array(
'id' => '5094',
'name' => 'Transient loss of Polycomb components induces an epigenetic cancer fate',
'authors' => 'V. Parreno et al.',
'description' => '<p><span>Although cancer initiation and progression are generally associated with the accumulation of somatic mutations</span><sup><a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 1" title="McGranahan, N. & Swanton, C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 27, 15–26 (2015)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR1" id="ref-link-section-d121858e719">1</a>,<a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 2" title="Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR2" id="ref-link-section-d121858e722">2</a></sup><span>, substantial epigenomic alterations underlie many aspects of tumorigenesis and cancer susceptibility</span><sup><a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" title="Brock, A., Chang, H. & Huang, S. Non-genetic heterogeneity—a mutation-independent driving force for the somatic evolution of tumours. Nat. Rev. Genet. 10, 336–342 (2009)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR3" id="ref-link-section-d121858e726">3</a>,<a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" title="Flavahan, W. A., Gaskell, E. & Bernstein, B. E. Epigenetic plasticity and the hallmarks of cancer. Science 357, eaal2380 (2017)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR4" id="ref-link-section-d121858e726_1">4</a>,<a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" title="Marine, J. C., Dawson, S. J. & Dawson, M. A. Non-genetic mechanisms of therapeutic resistance in cancer. Nat. Rev. Cancer 20, 743–756 (2020)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR5" id="ref-link-section-d121858e726_2">5</a>,<a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 6" title="Timp, W. & Feinberg, A. P. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat. Rev. Cancer 13, 497–510 (2013)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR6" id="ref-link-section-d121858e729">6</a></sup><span>, suggesting that genetic mechanisms might not be the only drivers of malignant transformation</span><sup><a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 7" title="Teixeira, V. H. et al. Deciphering the genomic, epigenomic, and transcriptomic landscapes of pre-invasive lung cancer lesions. Nat. Med. 25, 517–525 (2019)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR7" id="ref-link-section-d121858e733">7</a></sup><span>. However, whether purely non-genetic mechanisms are sufficient to initiate tumorigenesis irrespective of mutations has been unknown. Here, we show that a transient perturbation of transcriptional silencing mediated by Polycomb group proteins is sufficient to induce an irreversible switch to a cancer cell fate in<span> </span></span><i>Drosophila</i><span>. This is linked to the irreversible derepression of genes that can drive tumorigenesis, including members of the JAK–STAT signalling pathway and<span> </span></span><i>zfh1</i><span>, the fly homologue of the<span> </span></span><i>ZEB1</i><span><span> </span>oncogene, whose aberrant activation is required for Polycomb perturbation-induced tumorigenesis. These data show that a reversible depletion of Polycomb proteins can induce cancer in the absence of driver mutations, suggesting that tumours can emerge through epigenetic dysregulation leading to inheritance of altered cell fates.</span></p>',
'date' => '2024-04-24',
'pmid' => 'https://www.nature.com/articles/s41586-024-07328-w',
'doi' => 'https://doi.org/10.1038/s41586-024-07328-w',
'modified' => '2025-03-27 17:00:44',
'created' => '2025-03-27 17:00:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 15 => array(
'id' => '4933',
'name' => 'Systematic mapping of TF-mediated cell fate changes by a pooled induction coupled with scRNA-seq and multi-omics approaches',
'authors' => 'Lee M. et al.',
'description' => '<p><span>Transcriptional regulation controls cellular functions through interactions between transcription factors (TFs) and their chromosomal targets. However, understanding the fate conversion potential of multiple TFs in an inducible manner remains limited. Here, we introduce iTF-seq as a method for identifying individual TFs that can alter cell fate toward specific lineages at a single-cell level. iTF-seq enables time course monitoring of transcriptome changes, and with biotinylated individual TFs, it provides a multi-omics approach to understanding the mechanisms behind TF-mediated cell fate changes. Our iTF-seq study in mouse embryonic stem cells identified multiple TFs that trigger rapid transcriptome changes indicative of differentiation within a day of induction. Moreover, cells expressing these potent TFs often show a slower cell cycle and increased cell death. Further analysis using bioChIP-seq revealed that GCM1 and OTX2 act as pioneer factors and activators by increasing gene accessibility and activating the expression of lineage specification genes during cell fate conversion. iTF-seq has utility in both mapping cell fate conversion and understanding cell fate conversion mechanisms.</span></p>',
'date' => '2024-04-05',
'pmid' => 'https://pubmed.ncbi.nlm.nih.gov/38580401/',
'doi' => '10.1101/gr.277926.123',
'modified' => '2024-04-09 00:19:18',
'created' => '2024-04-09 00:19:18',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 16 => array(
'id' => '4895',
'name' => 'Protocol to isolate nuclei from Chlamydomonas reinhardtii for ATAC sequencing',
'authors' => 'Santhanagopalan I. et al.',
'description' => '<p class="section-title u-h4 u-margin-l-top u-margin-xs-bottom">Highlights</p>
<div id="abssec0020">
<ul class="list">
<li class="react-xocs-list-item">
<p id="p0010">Optimized isolation of nuclei from the green model alga<span> </span><em>Chlamydomonas reinhardtii</em></p>
</li>
<li class="react-xocs-list-item">
<p id="p0010"><em></em></p>
<p id="p0015">Tag-free isolation from both cell-walled and cell wall-deficient algae strains</p>
</li>
<li class="react-xocs-list-item">
<p id="p0015"></p>
<p id="p0020">Key steps for an effective and fast isolation and quantification procedure of nuclei</p>
</li>
<li class="react-xocs-list-item">
<p><span class="list-label"></span>Extracts at a quality suitable for ATAC-sequencing</p>
</li>
</ul>
</div>',
'date' => '2024-03-15',
'pmid' => 'https://www.sciencedirect.com/science/article/pii/S2666166723007311',
'doi' => 'https://doi.org/10.1016/j.xpro.2023.102764',
'modified' => '2024-02-09 12:37:32',
'created' => '2024-01-22 13:39:58',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 17 => array(
'id' => '4917',
'name' => 'Interplay between coding and non-coding regulation drives the Arabidopsis seed-to-seedling transition',
'authors' => 'Trembley B.J.M. et al.',
'description' => '<p><span>Translation of seed stored mRNAs is essential to trigger germination. However, when RNAPII re-engages RNA synthesis during the seed-to-seedling transition has remained in question. Combining csRNA-seq, ATAC-seq and smFISH in </span><i>Arabidopsis thaliana</i><span><span> </span>we demonstrate that active transcription initiation is detectable during the entire germination process. Features of non-coding regulation such as dynamic changes in chromatin accessible regions, antisense transcription, as well as bidirectional non-coding promoters are widespread throughout the Arabidopsis genome. We show that sensitivity to exogenous ABSCISIC ACID (ABA) during germination depends on proximal promoter accessibility at ABA-responsive genes. Moreover, we provide genetic validation of the existence of divergent transcription in plants. Our results reveal that active enhancer elements are transcribed producing non-coding enhancer RNAs (eRNAs) as widely documented in metazoans. In sum, this study defining the extent and role of coding and non-coding transcription during key stages of germination expands our understanding of transcriptional mechanisms underlying plant developmental transitions.</span></p>',
'date' => '2024-02-26',
'pmid' => 'https://www.nature.com/articles/s41467-024-46082-5',
'doi' => 'https://doi.org/10.1038/s41467-024-46082-5',
'modified' => '2024-02-29 11:56:56',
'created' => '2024-02-29 11:56:56',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 18 => array(
'id' => '4910',
'name' => 'The transcriptional regulatory network modulating human trophoblast stem cells to extravillous trophoblast differentiation',
'authors' => 'Kim M. et al.',
'description' => '<p><span>During human pregnancy, extravillous trophoblasts play crucial roles in placental invasion into the maternal decidua and spiral artery remodeling. However, regulatory factors and their action mechanisms modulating human extravillous trophoblast specification have been unknown. By analyzing dynamic changes in transcriptome and enhancer profile during human trophoblast stem cell to extravillous trophoblast differentiation, we define stage-specific regulators, including an early-stage transcription factor, TFAP2C, and multiple late-stage transcription factors. Loss-of-function studies confirm the requirement of all transcription factors identified for adequate differentiation, and we reveal that the dynamic changes in the levels of TFAP2C are essential. Notably, TFAP2C pre-occupies the regulatory elements of the inactive extravillous trophoblast-active genes during the early stage of differentiation, and the late-stage transcription factors directly activate extravillous trophoblast-active genes, including themselves as differentiation further progresses, suggesting sequential actions of transcription factors assuring differentiation. Our results reveal stage-specific transcription factors and their inter-connected regulatory mechanisms modulating extravillous trophoblast differentiation, providing a framework for understanding early human placentation and placenta-related complications.</span></p>',
'date' => '2024-02-12',
'pmid' => 'https://www.nature.com/articles/s41467-024-45669-2',
'doi' => 'https://doi.org/10.1038/s41467-024-45669-2',
'modified' => '2024-02-15 10:43:52',
'created' => '2024-02-15 10:43:52',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 19 => array(
'id' => '4928',
'name' => 'DMSO derives Trophectoderm and Clonal Blastoid from Single Human Pluripotent Stem Cell',
'authors' => 'Alsolami S. et al.',
'description' => '<p><span>Human naïve pluripotent stem cells (nPSCs) can differentiate into extra-embryonic trophectoderm (TE), a critical step in the generation of the integrated embryo model termed blastoid. The current paradigm of blastoid generation necessitates the aggregation of dozens of nPSCs treated with multiple small molecule inhibitors, growth factors, or genetic modifications to initiate TE differentiation. The presence of complex crosstalk among pathways and cellular heterogeneity in these models complicates mechanistic study and genetic screens. Here, we show that a single small molecule, dimethyl sulfoxide (DMSO), potently induces TE differentiation in basal medium without pharmacological and genetic perturbations. DMSO enhances blastoid generation and, more importantly, is sufficient for blastoid generation by itself. DMSO blastoids resemble blastocysts in morphology and lineage composition. DMSO induces blastoid formation through PKC signaling and cell cycle regulation. Lastly, DMSO enables single nPSC-derived clonal blastoids, which could facilitate genetic screens for mechanistic understanding of human embryogenesis.</span></p>',
'date' => '2024-01-01',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2023.12.31.573770v1.abstract',
'doi' => 'https://doi.org/10.1101/2023.12.31.573770',
'modified' => '2024-03-27 15:18:04',
'created' => '2024-03-27 15:18:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 20 => array(
'id' => '4887',
'name' => 'In vitro production of cat-restricted Toxoplasma pre-sexual stages',
'authors' => 'Antunes, A.V. et al.',
'description' => '<p><span>Sexual reproduction of </span><i>Toxoplasma gondii</i><span>, confined to the felid gut, remains largely uncharted owing to ethical concerns regarding the use of cats as model organisms. Chromatin modifiers dictate the developmental fate of the parasite during its multistage life cycle, but their targeting to stage-specific cistromes is poorly described</span><sup><a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 1" title="Farhat, D. C. et al. A MORC-driven transcriptional switch controls Toxoplasma developmental trajectories and sexual commitment. Nat. Microbiol. 5, 570–583 (2020)." href="https://www.nature.com/articles/s41586-023-06821-y#ref-CR1" id="ref-link-section-d277698175e527">1</a>,<a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 2" title="Bougdour, A. et al. Drug inhibition of HDAC3 and epigenetic control of differentiation in Apicomplexa parasites. J. Exp. Med. 206, 953–966 (2009)." href="https://www.nature.com/articles/s41586-023-06821-y#ref-CR2" id="ref-link-section-d277698175e530">2</a></sup><span>. Here we found that the transcription factors AP2XII-1 and AP2XI-2 operate during the tachyzoite stage, a hallmark of acute toxoplasmosis, to silence genes necessary for merozoites, a developmental stage critical for subsequent sexual commitment and transmission to the next host, including humans. Their conditional and simultaneous depletion leads to a marked change in the transcriptional program, promoting a full transition from tachyzoites to merozoites. These in vitro-cultured pre-gametes have unique protein markers and undergo typical asexual endopolygenic division cycles. In tachyzoites, AP2XII-1 and AP2XI-2 bind DNA as heterodimers at merozoite promoters and recruit MORC and HDAC3 (ref. </span><sup><a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 1" title="Farhat, D. C. et al. A MORC-driven transcriptional switch controls Toxoplasma developmental trajectories and sexual commitment. Nat. Microbiol. 5, 570–583 (2020)." href="https://www.nature.com/articles/s41586-023-06821-y#ref-CR1" id="ref-link-section-d277698175e534">1</a></sup><span>), thereby limiting chromatin accessibility and transcription. Consequently, the commitment to merogony stems from a profound epigenetic rewiring orchestrated by AP2XII-1 and AP2XI-2. Successful production of merozoites in vitro paves the way for future studies on<span> </span></span><i>Toxoplasma</i><span><span> </span>sexual development without the need for cat infections and holds promise for the development of therapies to prevent parasite transmission.</span></p>',
'date' => '2023-12-13',
'pmid' => 'https://www.nature.com/articles/s41586-023-06821-y',
'doi' => 'https://doi.org/10.1038/s41586-023-06821-y',
'modified' => '2023-12-18 10:40:50',
'created' => '2023-12-18 10:40:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 21 => array(
'id' => '4939',
'name' => 'Vaginal microbes alter epithelial transcriptomic and epigenomic modifications providing insight into the molecular mechanisms for susceptibility to adverse reproductive outcomes',
'authors' => 'Elovitz M. et al.',
'description' => '<p><span>The cervicovaginal microbiome is highly associated with women's health with microbial communities dominated by </span><i>Lactobacillus</i><span><span> </span>spp. being considered optimal. Conversely, a lack of lactobacilli and a high abundance of strict and facultative anaerobes including<span> </span></span><i>Gardnerella vaginalis</i><span>, have been associated with adverse reproductive outcomes. However, the molecular pathways modulated by microbe interactions with the cervicovaginal epithelia remain unclear. Using RNA-sequencing, we characterize the<span> </span></span><i>in vitro</i><span><span> </span>cervicovaginal epithelial transcriptional response to different vaginal bacteria and their culture supernatants. We showed that<span> </span></span><i>G. vaginalis</i><span><span> </span>upregulated genes were associated with an activated innate immune response including anti-microbial peptides and inflammasome pathways, represented by NLRP3-mediated increases in caspase-1, IL-1β and cell death. Cervicovaginal epithelial cells exposed to<span> </span></span><i>L. crispatus</i><span><span> </span>showed limited transcriptomic changes, while exposure to<span> </span></span><i>L. crispatus</i><span><span> </span>culture supernatants resulted in a shift in the epigenomic landscape of cervical epithelial cells. ATAC-sequencing confirmed epigenetic changes with reduced chromatin accessibility. This study reveals new insight into host-microbe interactions in the lower reproductive tract and suggest potential therapeutic strategies leveraging the vaginal microbiome to improve reproductive health.</span></p>',
'date' => '2023-11-16',
'pmid' => 'https://pubmed.ncbi.nlm.nih.gov/38014044/',
'doi' => '10.21203/rs.3.rs-3580132/v1',
'modified' => '2024-06-07 15:47:48',
'created' => '2024-06-07 15:47:48',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 22 => array(
'id' => '4927',
'name' => 'Inflammatory stress-mediated chromatin changes underlie dysfunction in endothelial cells',
'authors' => 'Liu H. et al.',
'description' => '<p><span>Inflammatory stresses underlie endothelial dysfunction and contribute to the development of chronic cardiovascular disorders such as atherosclerosis and vascular fibrosis. The initial transcriptional response of endothelial cells to pro-inflammatory cytokines such as TNF-alpha is well established. However, very few studies uncover the effects of inflammatory stresses on chromatin architecture. We used integrative analysis of ATAC-seq and RNA-seq data to investigate chromatin alterations in human endothelial cells in response to TNF-alpha and febrile-range heat stress exposure. Multi-omics data analysis suggests a correlation between the transcription of stress-related genes and endothelial dysfunction drivers with chromatin regions exhibiting differential accessibility. Moreover, microscopy identified the dynamics in the nuclear organization, specifically, the changes in a subset of heterochromatic nucleoli-associated chromatin domains, the centromeres. Upon inflammatory stress exposure, the centromeres decreased association with nucleoli in a p38-dependent manner and increased the number of transcripts from pericentromeric regions. Overall, we provide two lines of evidence that suggest chromatin alterations in vascular endothelial cells during inflammatory stresses.</span></p>',
'date' => '2023-10-16',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10614786/',
'doi' => '10.1101/2023.10.11.561959',
'modified' => '2024-03-27 15:15:21',
'created' => '2024-03-27 15:15:21',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 23 => array(
'id' => '4880',
'name' => 'Dynamic PRC1-CBX8 stabilizes a porous structure of chromatin condensates',
'authors' => 'Uckelmann M. et al.',
'description' => '<p><span>The compaction of chromatin is a prevalent paradigm in gene repression. Chromatin compaction is commonly thought to repress transcription by restricting chromatin accessibility. However, the spatial organisation and dynamics of chromatin compacted by gene-repressing factors are unknown. Using cryo-electron tomography, we solved the three dimensional structure of chromatin condensed by the Polycomb Repressive Complex 1 (PRC1) in a complex with CBX8. PRC1-condensed chromatin is porous and stabilised through multivalent dynamic interactions of PRC1 with chromatin. Within condensates, PRC1 remains dynamic while maintaining a static chromatin structure. In differentiated mouse embryonic stem cells, CBX8-bound chromatin remains accessible. These findings challenge the idea of rigidly compacted polycomb domains and instead provides a mechanistic framework for dynamic and accessible PRC1-chromatin condensates.</span></p>',
'date' => '2023-05-09',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2023.05.08.539931v1.abstract',
'doi' => 'https://doi.org/10.1101/2023.05.08.539931',
'modified' => '2023-11-10 15:43:37',
'created' => '2023-11-10 15:43:37',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 24 => array(
'id' => '4810',
'name' => 'UMSBP2 is chromatin remodeler that functions in regulation of geneexpression and suppression of antigenic variation in trypanosomes.',
'authors' => 'Soni A. et al.',
'description' => '<p><span>Universal Minicircle Sequence binding proteins (UMSBPs) are CCHC-type zinc-finger proteins that bind the single-stranded G-rich UMS sequence, conserved at the replication origins of minicircles in the kinetoplast DNA, the mitochondrial genome of kinetoplastids. Trypanosoma brucei UMSBP2 has been recently shown to colocalize with telomeres and to play an essential role in chromosome end protection. Here we report that TbUMSBP2 decondenses in vitro DNA molecules, which were condensed by core histones H2B, H4 or linker histone H1. DNA decondensation is mediated via protein-protein interactions between TbUMSBP2 and these histones, independently of its previously described DNA binding activity. Silencing of the TbUMSBP2 gene resulted in a significant decrease in the disassembly of nucleosomes in T. brucei chromatin, a phenotype that could be reverted, by supplementing the knockdown cells with TbUMSBP2. Transcriptome analysis revealed that silencing of TbUMSBP2 affects the expression of multiple genes in T. brucei, with a most significant effect on the upregulation of the subtelomeric variant surface glycoproteins (VSG) genes, which mediate the antigenic variation in African trypanosomes. These observations suggest that UMSBP2 is a chromatin remodeling protein that functions in the regulation of gene expression and plays a role in the control of antigenic variation in T. brucei.</span></p>',
'date' => '2023-05-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/37207337',
'doi' => '10.1093/nar/gkad402',
'modified' => '2023-06-15 08:54:17',
'created' => '2023-06-13 21:11:31',
'ProductsPublication' => array(
[maximum depth reached]
)
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
'id' => '2440',
'name' => 'ATAC-seq kit SDS US en',
'language' => 'en',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-US-en-1_0.pdf',
'countries' => 'US',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '2438',
'name' => 'ATAC-seq kit SDS GB en',
'language' => 'en',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-GB-en-1_0.pdf',
'countries' => 'GB',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '2437',
'name' => 'ATAC-seq kit SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-FR-fr-1_0.pdf',
'countries' => 'FR',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '2433',
'name' => 'ATAC-seq kit SDS BE fr',
'language' => 'fr',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-BE-fr-1_0.pdf',
'countries' => 'BE',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '2436',
'name' => 'ATAC-seq kit SDS ES es',
'language' => 'es',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-ES-es-1_0.pdf',
'countries' => 'ES',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '2439',
'name' => 'ATAC-seq kit SDS JP ja',
'language' => 'ja',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-JP-ja-1_0.pdf',
'countries' => 'JP',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '2435',
'name' => 'ATAC-seq kit SDS DE de',
'language' => 'de',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-DE-de-1_0.pdf',
'countries' => 'DE',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '2434',
'name' => 'ATAC-seq kit SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-BE-nl-1_0.pdf',
'countries' => 'BE',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
)
)
)
$meta_canonical = 'https://dev.diagenode.com/en/p/atac-seq-kit-24rxns'
$fromEmail = 'dev.diagenode.com'
$country = 'US'
$countries_allowed = array(
(int) 0 => 'CA',
(int) 1 => 'US',
(int) 2 => 'IE',
(int) 3 => 'GB',
(int) 4 => 'DK',
(int) 5 => 'NO',
(int) 6 => 'SE',
(int) 7 => 'FI',
(int) 8 => 'NL',
(int) 9 => 'BE',
(int) 10 => 'LU',
(int) 11 => 'FR',
(int) 12 => 'DE',
(int) 13 => 'CH',
(int) 14 => 'AT',
(int) 15 => 'ES',
(int) 16 => 'IT',
(int) 17 => 'PT'
)
$outsource = false
$other_formats = array(
(int) 0 => array(
'id' => '3193',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<p><a href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p><strong>ATAC-seq</strong>, Assay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the <strong>transposase Tn5</strong> which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
<p>The Diagenode’s <b>ATAC-</b><b>seq</b><b> kit </b>is based on a highly validated protocol optimized for <b>50,000 </b><b>cells</b><b> per </b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The primer indexes for multiplexing are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell </b><b>requirement</b><b>: </b><b>50,000 </b><b>cells / </b><b>rxn</b></li>
<li><b>Robust protocol </b>with <b>high reproducibility </b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong> and <b>efficient DNA capture </b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids </b><b>over-amplification </b></li>
<li>Allows adaptation/flexibility for <b>more challenging samples </b>to succeed with library prep.</li>
<li>Gives <strong>early indication</strong> if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p><span>Looking for ATAC-seq on tissue? Please, go to: </span><a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack: <a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"> 0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"> (Tn5 transposase) </a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"> Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '8 rxns',
'catalog_number' => 'C01080001',
'old_catalog_number' => '',
'sf_code' => 'C01080001-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '615',
'price_USD' => '615',
'price_GBP' => '535',
'price_JPY' => '110700',
'price_CNY' => '',
'price_AUD' => '1538',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-8rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment (C01080001)| Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2023-05-05 07:57:08',
'created' => '2021-06-24 16:47:47',
'ProductsGroup' => array(
'id' => '397',
'product_id' => '3193',
'group_id' => '349'
)
)
)
$pro = array(
'id' => '3193',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<p><a href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p><strong>ATAC-seq</strong>, Assay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the <strong>transposase Tn5</strong> which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
<p>The Diagenode’s <b>ATAC-</b><b>seq</b><b> kit </b>is based on a highly validated protocol optimized for <b>50,000 </b><b>cells</b><b> per </b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The primer indexes for multiplexing are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell </b><b>requirement</b><b>: </b><b>50,000 </b><b>cells / </b><b>rxn</b></li>
<li><b>Robust protocol </b>with <b>high reproducibility </b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong> and <b>efficient DNA capture </b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids </b><b>over-amplification </b></li>
<li>Allows adaptation/flexibility for <b>more challenging samples </b>to succeed with library prep.</li>
<li>Gives <strong>early indication</strong> if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p><span>Looking for ATAC-seq on tissue? Please, go to: </span><a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack: <a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"> 0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"> (Tn5 transposase) </a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"> Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '8 rxns',
'catalog_number' => 'C01080001',
'old_catalog_number' => '',
'sf_code' => 'C01080001-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '615',
'price_USD' => '615',
'price_GBP' => '535',
'price_JPY' => '110700',
'price_CNY' => '',
'price_AUD' => '1538',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-8rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment (C01080001)| Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2023-05-05 07:57:08',
'created' => '2021-06-24 16:47:47',
'ProductsGroup' => array(
'id' => '397',
'product_id' => '3193',
'group_id' => '349'
)
)
$edit = ''
$testimonials = ''
$featured_testimonials = ''
$related_products = ''
$rrbs_service = array(
(int) 0 => (int) 1894,
(int) 1 => (int) 1895
)
$chipseq_service = array(
(int) 0 => (int) 2683,
(int) 1 => (int) 1835,
(int) 2 => (int) 1836,
(int) 3 => (int) 2684,
(int) 4 => (int) 1838,
(int) 5 => (int) 1839,
(int) 6 => (int) 1856
)
$labelize = object(Closure) {
}
$old_catalog_number = ''
$country_code = 'US'
$other_format = array(
'id' => '3193',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<p><a href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p><strong>ATAC-seq</strong>, Assay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the <strong>transposase Tn5</strong> which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
<p>The Diagenode’s <b>ATAC-</b><b>seq</b><b> kit </b>is based on a highly validated protocol optimized for <b>50,000 </b><b>cells</b><b> per </b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The primer indexes for multiplexing are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell </b><b>requirement</b><b>: </b><b>50,000 </b><b>cells / </b><b>rxn</b></li>
<li><b>Robust protocol </b>with <b>high reproducibility </b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong> and <b>efficient DNA capture </b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids </b><b>over-amplification </b></li>
<li>Allows adaptation/flexibility for <b>more challenging samples </b>to succeed with library prep.</li>
<li>Gives <strong>early indication</strong> if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p><span>Looking for ATAC-seq on tissue? Please, go to: </span><a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack: <a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"> 0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"> (Tn5 transposase) </a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"> Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '8 rxns',
'catalog_number' => 'C01080001',
'old_catalog_number' => '',
'sf_code' => 'C01080001-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '615',
'price_USD' => '615',
'price_GBP' => '535',
'price_JPY' => '110700',
'price_CNY' => '',
'price_AUD' => '1538',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-8rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment (C01080001)| Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2023-05-05 07:57:08',
'created' => '2021-06-24 16:47:47',
'ProductsGroup' => array(
'id' => '397',
'product_id' => '3193',
'group_id' => '349'
)
)
$label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>'
$document = array(
'id' => '1139',
'name' => 'ATAC-seq Kit - Manual',
'description' => '<p class="p1">Gene expression is carefully regulated in the cells in order to manage a wide range of biological functions. The structure of chromatin is quite dynamic and contributes to this crucial regulatory process.</p>
<p class="p1">ATAC-seq, Assay for Transposase-Accessible Chromatin, followed by nextgeneration sequencing, is a key technology to easily identify the “open” regions of the chromatin, which are usually associated with permissive gene expression. Indeed, the nuclei of the samples are incubated with a transposase, and only the genomic regions associated with open chromatin will be accessible to this transposase. During the process those regions will be cut and sequencing adaptors will be added, allowing their sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only., giving a map of the chromatin status in the whole genome of the sample.</p>
<p class="p1">The Diagenode’s ATAC-seq kit is based on a highly validated protocol, used for years in our Epigenomics Profiling Services offer and takes advantage of many successful Diagenode’s tools, such as the loaded Tagmentase (Tn5 transposase), the MicroChIP DiaPure Columns and the Primer indexes for tagmented libraries kits.</p>',
'image_id' => null,
'type' => 'Manual',
'url' => 'files/products/kits/atacseq-kit-manual.pdf',
'slug' => 'atacseq-kit-manual',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-06-25 09:51:59',
'created' => '2021-06-25 09:51:59',
'ProductsDocument' => array(
'id' => '3130',
'product_id' => '3194',
'document_id' => '1139'
)
)
$sds = array(
'id' => '2434',
'name' => 'ATAC-seq kit SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-BE-nl-1_0.pdf',
'countries' => 'BE',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
'id' => '4140',
'product_id' => '3194',
'safety_sheet_id' => '2434'
)
)
$publication = array(
'id' => '4810',
'name' => 'UMSBP2 is chromatin remodeler that functions in regulation of geneexpression and suppression of antigenic variation in trypanosomes.',
'authors' => 'Soni A. et al.',
'description' => '<p><span>Universal Minicircle Sequence binding proteins (UMSBPs) are CCHC-type zinc-finger proteins that bind the single-stranded G-rich UMS sequence, conserved at the replication origins of minicircles in the kinetoplast DNA, the mitochondrial genome of kinetoplastids. Trypanosoma brucei UMSBP2 has been recently shown to colocalize with telomeres and to play an essential role in chromosome end protection. Here we report that TbUMSBP2 decondenses in vitro DNA molecules, which were condensed by core histones H2B, H4 or linker histone H1. DNA decondensation is mediated via protein-protein interactions between TbUMSBP2 and these histones, independently of its previously described DNA binding activity. Silencing of the TbUMSBP2 gene resulted in a significant decrease in the disassembly of nucleosomes in T. brucei chromatin, a phenotype that could be reverted, by supplementing the knockdown cells with TbUMSBP2. Transcriptome analysis revealed that silencing of TbUMSBP2 affects the expression of multiple genes in T. brucei, with a most significant effect on the upregulation of the subtelomeric variant surface glycoproteins (VSG) genes, which mediate the antigenic variation in African trypanosomes. These observations suggest that UMSBP2 is a chromatin remodeling protein that functions in the regulation of gene expression and plays a role in the control of antigenic variation in T. brucei.</span></p>',
'date' => '2023-05-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/37207337',
'doi' => '10.1093/nar/gkad402',
'modified' => '2023-06-15 08:54:17',
'created' => '2023-06-13 21:11:31',
'ProductsPublication' => array(
'id' => '6971',
'product_id' => '3194',
'publication_id' => '4810'
)
)
$externalLink = ' <a href="https://www.ncbi.nlm.nih.gov/pubmed/37207337" target="_blank"><i class="fa fa-external-link"></i></a>'CustomErrorHandler::handleError() - APP/Lib/CustomErrorHandler.php, line 9
include - APP/View/Products/view.ctp, line 755
View::_evaluate() - CORE/Cake/View/View.php, line 971
View::_render() - CORE/Cake/View/View.php, line 933
View::render() - CORE/Cake/View/View.php, line 473
Controller::render() - CORE/Cake/Controller/Controller.php, line 963
ProductsController::slug() - APP/Controller/ProductsController.php, line 1055
ReflectionMethod::invokeArgs() - [internal], line ??
Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491
Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193
Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167
[main] - APP/webroot/index.php, line 118
(Notice): Undefined variable: campaign_id [APP/View/Products/view.ctp, line 755]Code Context $errorType = self::getErrorType($code);
self::logToDatabase( $errorType, $description, $file, $line);
return parent::handleError( $errorType, $description, $file, $line, $context);
$viewFile = '/var/www/dev.diagenode.com/app/View/Products/view.ctp'
$dataForView = array(
'language' => 'en',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'meta_title' => 'ATAC-seq kit for open chromatin assessment C01080001 | Diagenode ',
'product' => array(
'Product' => array(
'id' => '3194',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<div class="row">
<div class="small-12 medium-8 large-8 columns">
<p><a class="tiny details button" href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf">Download the Manual</a></p>
<p><strong>ATAC-seq</strong>, Assay for<span> </span><strong>T</strong>ransposase-<strong>A</strong>ccessible<span> </span><strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the<span> </span><strong>transposase Tn5</strong><span> </span>which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
</div>
<div class="small-12 medium-4 large-4 columns"><center>
<script>// <![CDATA[
var date = new Date(); var heure = date.getHours(); var jour = date.getDay(); var semaine = Math.floor(date.getDate() / 7) + 1; if (jour === 2 && ( (heure >= 9 && heure < 9.5) || (heure >= 18 && heure < 18.5) )) { document.write('<a href="https://us02web.zoom.us/j/85467619762"><img src="https://www.diagenode.com/img/epicafe-ON.gif"></a>'); } else { document.write('<a href="https://go.diagenode.com/l/928883/2023-04-26/3kq1v"><img src="https://www.diagenode.com/img/epicafe-OFF.png"></a>'); }
// ]]></script>
</center></div>
</div>
<p>Diagenode’s<span> </span><b>ATAC-</b><b>seq</b><b><span> </span>kit<span> </span></b>is based on a highly validated protocol optimized for<span> </span><b>50,000<span> </span></b><b>cells</b><b><span> </span>per<span> </span></b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The <a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">primer indexes for multiplexing</a> are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell<span> </span></b><b>requirement</b><b>:<span> </span></b><b>50,000<span> </span></b><b>cells /<span> </span></b><b>rxn</b></li>
<li><b>Robust protocol<span> </span></b>with<span> </span><b>high reproducibility<span> </span></b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong><span> </span>and<span> </span><b>efficient DNA capture<span> </span></b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids<span> </span></b><b>over-amplification</b></li>
<li>Allows adaptation/flexibility for<span> </span><b>more challenging samples<span> </span></b>to succeed with library prep.</li>
<li>Gives<span> </span><strong>early indication</strong><span> </span>if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p>Looking for ATAC-seq on tissue? Please, go to: <a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack:<span> </span><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"><span> </span>0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"><span> </span>(Tn5 transposase)<span> </span></a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"><span> </span>Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '24 rxns',
'catalog_number' => 'C01080002',
'old_catalog_number' => '',
'sf_code' => 'C01080002-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '1740',
'price_USD' => '1745',
'price_GBP' => '1485',
'price_JPY' => '313200',
'price_CNY' => '',
'price_AUD' => '4362',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-24rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment C01080001 | Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2025-04-17 10:29:58',
'created' => '2021-07-01 15:46:58',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => null,
'name' => null,
'description' => null,
'clonality' => null,
'isotype' => null,
'lot' => null,
'concentration' => null,
'reactivity' => null,
'type' => null,
'purity' => null,
'classification' => null,
'application_table' => null,
'storage_conditions' => null,
'storage_buffer' => null,
'precautions' => null,
'uniprot_acc' => null,
'slug' => null,
'meta_keywords' => null,
'meta_description' => null,
'modified' => null,
'created' => null,
'select_label' => null
),
'Slave' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Group' => array(
'Group' => array(
[maximum depth reached]
),
'Master' => array(
[maximum depth reached]
),
'Product' => array(
[maximum depth reached]
)
),
'Related' => array(),
'Application' => array(),
'Category' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Document' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
[maximum depth reached]
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
),
(int) 8 => array(
[maximum depth reached]
),
(int) 9 => array(
[maximum depth reached]
),
(int) 10 => array(
[maximum depth reached]
),
(int) 11 => array(
[maximum depth reached]
),
(int) 12 => array(
[maximum depth reached]
),
(int) 13 => array(
[maximum depth reached]
),
(int) 14 => array(
[maximum depth reached]
),
(int) 15 => array(
[maximum depth reached]
),
(int) 16 => array(
[maximum depth reached]
),
(int) 17 => array(
[maximum depth reached]
),
(int) 18 => array(
[maximum depth reached]
),
(int) 19 => array(
[maximum depth reached]
),
(int) 20 => array(
[maximum depth reached]
),
(int) 21 => array(
[maximum depth reached]
),
(int) 22 => array(
[maximum depth reached]
),
(int) 23 => array(
[maximum depth reached]
),
(int) 24 => array(
[maximum depth reached]
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
[maximum depth reached]
),
(int) 1 => array(
[maximum depth reached]
),
(int) 2 => array(
[maximum depth reached]
),
(int) 3 => array(
[maximum depth reached]
),
(int) 4 => array(
[maximum depth reached]
),
(int) 5 => array(
[maximum depth reached]
),
(int) 6 => array(
[maximum depth reached]
),
(int) 7 => array(
[maximum depth reached]
)
)
),
'meta_canonical' => 'https://dev.diagenode.com/en/p/atac-seq-kit-24rxns',
'fromEmail' => 'dev.diagenode.com'
)
$language = 'en'
$meta_keywords = ''
$meta_description = 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility'
$meta_title = 'ATAC-seq kit for open chromatin assessment C01080001 | Diagenode '
$product = array(
'Product' => array(
'id' => '3194',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<div class="row">
<div class="small-12 medium-8 large-8 columns">
<p><a class="tiny details button" href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf">Download the Manual</a></p>
<p><strong>ATAC-seq</strong>, Assay for<span> </span><strong>T</strong>ransposase-<strong>A</strong>ccessible<span> </span><strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the<span> </span><strong>transposase Tn5</strong><span> </span>which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
</div>
<div class="small-12 medium-4 large-4 columns"><center>
<script>// <![CDATA[
var date = new Date(); var heure = date.getHours(); var jour = date.getDay(); var semaine = Math.floor(date.getDate() / 7) + 1; if (jour === 2 && ( (heure >= 9 && heure < 9.5) || (heure >= 18 && heure < 18.5) )) { document.write('<a href="https://us02web.zoom.us/j/85467619762"><img src="https://www.diagenode.com/img/epicafe-ON.gif"></a>'); } else { document.write('<a href="https://go.diagenode.com/l/928883/2023-04-26/3kq1v"><img src="https://www.diagenode.com/img/epicafe-OFF.png"></a>'); }
// ]]></script>
</center></div>
</div>
<p>Diagenode’s<span> </span><b>ATAC-</b><b>seq</b><b><span> </span>kit<span> </span></b>is based on a highly validated protocol optimized for<span> </span><b>50,000<span> </span></b><b>cells</b><b><span> </span>per<span> </span></b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The <a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">primer indexes for multiplexing</a> are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell<span> </span></b><b>requirement</b><b>:<span> </span></b><b>50,000<span> </span></b><b>cells /<span> </span></b><b>rxn</b></li>
<li><b>Robust protocol<span> </span></b>with<span> </span><b>high reproducibility<span> </span></b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong><span> </span>and<span> </span><b>efficient DNA capture<span> </span></b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids<span> </span></b><b>over-amplification</b></li>
<li>Allows adaptation/flexibility for<span> </span><b>more challenging samples<span> </span></b>to succeed with library prep.</li>
<li>Gives<span> </span><strong>early indication</strong><span> </span>if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p>Looking for ATAC-seq on tissue? Please, go to: <a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack:<span> </span><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"><span> </span>0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"><span> </span>(Tn5 transposase)<span> </span></a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"><span> </span>Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '24 rxns',
'catalog_number' => 'C01080002',
'old_catalog_number' => '',
'sf_code' => 'C01080002-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '1740',
'price_USD' => '1745',
'price_GBP' => '1485',
'price_JPY' => '313200',
'price_CNY' => '',
'price_AUD' => '4362',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-24rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment C01080001 | Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2025-04-17 10:29:58',
'created' => '2021-07-01 15:46:58',
'locale' => 'eng'
),
'Antibody' => array(
'host' => '*****',
'id' => null,
'name' => null,
'description' => null,
'clonality' => null,
'isotype' => null,
'lot' => null,
'concentration' => null,
'reactivity' => null,
'type' => null,
'purity' => null,
'classification' => null,
'application_table' => null,
'storage_conditions' => null,
'storage_buffer' => null,
'precautions' => null,
'uniprot_acc' => null,
'slug' => null,
'meta_keywords' => null,
'meta_description' => null,
'modified' => null,
'created' => null,
'select_label' => null
),
'Slave' => array(
(int) 0 => array(
'id' => '349',
'name' => 'C01080002',
'product_id' => '3194',
'modified' => '2021-07-01 15:47:50',
'created' => '2021-07-01 15:47:50'
)
),
'Group' => array(
'Group' => array(
'id' => '349',
'name' => 'C01080002',
'product_id' => '3194',
'modified' => '2021-07-01 15:47:50',
'created' => '2021-07-01 15:47:50'
),
'Master' => array(
'id' => '3194',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<div class="row">
<div class="small-12 medium-8 large-8 columns">
<p><a class="tiny details button" href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf">Download the Manual</a></p>
<p><strong>ATAC-seq</strong>, Assay for<span> </span><strong>T</strong>ransposase-<strong>A</strong>ccessible<span> </span><strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the<span> </span><strong>transposase Tn5</strong><span> </span>which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
</div>
<div class="small-12 medium-4 large-4 columns"><center>
<script>// <![CDATA[
var date = new Date(); var heure = date.getHours(); var jour = date.getDay(); var semaine = Math.floor(date.getDate() / 7) + 1; if (jour === 2 && ( (heure >= 9 && heure < 9.5) || (heure >= 18 && heure < 18.5) )) { document.write('<a href="https://us02web.zoom.us/j/85467619762"><img src="https://www.diagenode.com/img/epicafe-ON.gif"></a>'); } else { document.write('<a href="https://go.diagenode.com/l/928883/2023-04-26/3kq1v"><img src="https://www.diagenode.com/img/epicafe-OFF.png"></a>'); }
// ]]></script>
</center></div>
</div>
<p>Diagenode’s<span> </span><b>ATAC-</b><b>seq</b><b><span> </span>kit<span> </span></b>is based on a highly validated protocol optimized for<span> </span><b>50,000<span> </span></b><b>cells</b><b><span> </span>per<span> </span></b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The <a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">primer indexes for multiplexing</a> are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell<span> </span></b><b>requirement</b><b>:<span> </span></b><b>50,000<span> </span></b><b>cells /<span> </span></b><b>rxn</b></li>
<li><b>Robust protocol<span> </span></b>with<span> </span><b>high reproducibility<span> </span></b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong><span> </span>and<span> </span><b>efficient DNA capture<span> </span></b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids<span> </span></b><b>over-amplification</b></li>
<li>Allows adaptation/flexibility for<span> </span><b>more challenging samples<span> </span></b>to succeed with library prep.</li>
<li>Gives<span> </span><strong>early indication</strong><span> </span>if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p>Looking for ATAC-seq on tissue? Please, go to: <a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack:<span> </span><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"><span> </span>0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"><span> </span>(Tn5 transposase)<span> </span></a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"><span> </span>Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a><span> </span><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '24 rxns',
'catalog_number' => 'C01080002',
'old_catalog_number' => '',
'sf_code' => 'C01080002-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '1740',
'price_USD' => '1745',
'price_GBP' => '1485',
'price_JPY' => '313200',
'price_CNY' => '',
'price_AUD' => '4362',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => true,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-24rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment C01080001 | Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2025-04-17 10:29:58',
'created' => '2021-07-01 15:46:58'
),
'Product' => array(
(int) 0 => array(
[maximum depth reached]
)
)
),
'Related' => array(),
'Application' => array(),
'Category' => array(
(int) 0 => array(
'id' => '13',
'position' => '2',
'parent_id' => null,
'name' => 'Chromatin studies',
'description' => '<div class="row">
<div class="small-12 medium-8 large-8 columns"><br />
<p>Chromatin structure plays a key role in regulating gene expression by allowing DNA accessibility to transcriptional machinery and transcription factors. The packaging of DNA into nucleosomes forms a closed structure that is not highly accessible to transcriptional elements whereas the open nucleosome structure allows DNA to be accessible. Diagenode offers a number of solutions to help you analyze chromatin and the role of transcriptional machinery including ChIP kits, ChIPmentation kits, antibodies, pA-Tn5 and ATAC-seq kits.</p>
</div>
<div class="small-12 medium-4 large-4 columns"><center>
<script>// <![CDATA[
var date = new Date(); var heure = date.getHours(); var jour = date.getDay(); var semaine = Math.floor(date.getDate() / 7) + 1; if (jour === 2 && ( (heure >= 9 && heure < 9.5) || (heure >= 18 && heure < 18.5) )) { document.write('<a href="https://us02web.zoom.us/j/85467619762"><img src="https://www.diagenode.com/img/epicafe-ON.gif"></a>'); } else { document.write('<a href="https://go.diagenode.com/l/928883/2023-04-26/3kq1v"><img src="https://www.diagenode.com/img/epicafe-OFF.png"></a>'); }
// ]]></script>
</center></div>
</div>
<p><a href="https://www.diagenode.com/en/p/uchipmentation-for-histones-24-rxns"><img src="https://www.diagenode.com/img/banners/b-microchip-category.png" /></a></p>
<div class="row">
<div class="small-12 medium-12 large-12 columns">
<div id="portal" class="main-portal">
<div class="portal-inner"><nav class="portal-nav">
<ul class="tips-menu">
<li><a href="#workflow" class="tips portal button" style="background: #13b29c; color: #f3fbfa;">Chromatin immunoprecipitation</a></li>
<li><a href="https://www.diagenode.com/en/categories/chromatin-ip-chipmentation" class="tips portal button">ChIPmentation</a></li>
<li><a href="https://www.diagenode.com/en/categories/antibodies
" class="tips portal button">Antibodies</a></li>
<li><a href="https://www.diagenode.com/en/categories/cutandtag" class="tips portal button">pA-Tn5</a></li>
<li><a href="https://www.diagenode.com/en/categories/atac-seq" class="tips portal button">ATAC-seq</a></li>
<li><a href="https://www.diagenode.com/en/categories/cutandtag" class="tips portal button">CUT&Tag</a></li>
</ul>
</nav></div>
</div>
<p>Chromatin immunoprecipitation (ChIP) determines the location of DNA binding sites on the genome for a protein of interest, giving insights into gene expression regulation. ChIP involves the selective enrichment of a chromatin fraction containing a specific antigen. Antibodies that recognize a protein or protein modification are used to determine the relative abundance of that antigen at a specific locus or loci. ChIP can be used to compare enrichment of proteins, map protein modifications, or quantify a protein modification during a time course.</p>
<span class="anchor" id="workflow"></span>
<h4><span style="font-weight: 400;">The ChIP workflow</span></h4>
<ul class="accordion" data-accordion="">
<li class="accordion-navigation"><img src="https://www.diagenode.com/img/categories/kits_chromatin_function/website-chip-workflow.jpg" />
<div id="chip_workflow" class="content">
<div class="row">
<table>
<tbody>
<tr>
<td width="50%">
<h3 class="text-center">Step by step workflow</h3>
</td>
<td width="50%">
<h3 class="text-center">Optimal solution from Diagenode</h3>
</td>
</tr>
<tr>
<td colspan="2"></td>
</tr>
<tr valign="top">
<td>
<p class="lead text-center"><strong>1.</strong><span> </span>Crosslink to bind proteins to DNA</p>
<center><img src="https://www.diagenode.com/img/chip/step-by-step-1-workflow.png" width="192" height="24" /></center></td>
<td>
<ul class="arrow">
<li><a href="https://www.diagenode.com/p/chip-cross-link-gold-600-ul">ChIP Cross-link Gold</a></li>
</ul>
</td>
</tr>
<tr>
<td colspan="2"></td>
</tr>
<tr valign="top">
<td>
<p class="lead text-center"><strong>2.</strong><span> </span>Shear DNA</p>
<center><img src="https://www.diagenode.com/img/chip/step-by-step-2-workflow.png" width="194" height="80" /></center></td>
<td>
<ul class="arrow">
<li><a href="https://www.diagenode.com/p/bioruptor-pico-sonication-device#">Bioruptor<sup>®</sup><span> </span>Pico Sonication device</a></li>
<li><a href="https://www.diagenode.com/categories/chromatin-shearing">Shearing optimization reagent</a></li>
</ul>
</td>
</tr>
<tr>
<td colspan="2"></td>
</tr>
<tr valign="top">
<td>
<p class="lead text-center"><strong>3.</strong><span> </span>Immunoprecipitate with specific antibody</p>
<center><img src="https://www.diagenode.com/img/chip/step-by-step-3-workflow.png" width="47" height="51" /></center></td>
<td>
<ul class="arrow">
<li><a href="https://www.diagenode.com/categories/chromatin-immunoprecipitation">ChIP-seq and ChIP-qPCR kits</a><span> </span>for transcription factors, histones, low inputs, plants</li>
<li><a href="https://www.diagenode.com/categories/chip-grade-antibodies">ChIP</a><span> </span>and<span> </span><a href="https://www.diagenode.com/categories/chip-seq-grade-antibodies">ChIP-seq grade</a><span> </span>antibodies</li>
<li><a href="https://www.diagenode.com/categories/ip-star">Automation available</a></li>
</ul>
</td>
</tr>
<tr>
<td colspan="2"></td>
</tr>
<tr valign="top">
<td>
<p class="lead text-center"><strong>4.</strong><span> </span>Reverse crosslinks and purify</p>
<center><img src="https://www.diagenode.com/img/chip/step-by-step-4-workflow.png" width="194" height="80" /></center></td>
<td>
<ul class="arrow">
<li>Diagenode's ChIP kits (contain optimal purification modules)</li>
</ul>
</td>
</tr>
<tr>
<td colspan="2"></td>
</tr>
<tr valign="top">
<td>
<p class="lead text-center"><strong>5.</strong><span> </span><a href="https://www.diagenode.com/en/p/ideal-chip-qpcr-kit">ChIP-qPCR</a><span> </span>or ChIP-seq library preparation</p>
</td>
<td>
<ul class="arrow">
<li>ChIP-seq:</li>
</ul>
<ul style="list-style-type: square;">
<li style="margin-left: 40px; font-size: 1.09rem;"><a href="https://www.diagenode.com/p/microplex-library-preparation-kit-v2-x48-12-indices-48-rxns">MicroPlex Library Preparation for 50 pg - 5 ng</a></li>
<li style="margin-left: 40px; font-size: 1.09rem;"><a href="https://www.diagenode.com/p/ideal-library-preparation-kit-x24-incl-index-primer-set-1-24-rxns">iDeal Library Preparation for > 5 ng</a></li>
</ul>
<ul class="arrow">
<li>ChIP qPCR</li>
</ul>
</td>
</tr>
</tbody>
</table>
</div>
</div>
</li>
</ul>
<p></p>
<h4><span style="font-weight: 400;">Products for chromatin study</span></h4>
<p><span style="font-weight: 400;"></span></p>
<p style="padding-left: 30px;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/crosslinking.png" alt="" width="35" height="35" /> Crosslinking<br /></b></span></strong><span style="font-weight: 400;">Efficient solution for protein-protein fixation.</span><strong><span style="font-weight: 400;"><b><span style="font-weight: 400;"> <a href="../p/chip-cross-link-gold-600-ul
">Read more</a></span></b><br /></span></strong></p>
<p style="padding-left: 30px;"><strong><img src="https://www.diagenode.com/img/applications/chromatin-shearing.png" alt="" width="35" height="35" /> Chromatin shearing<br /></strong><strong><span style="font-weight: 400;">Perfectly sheared chromatin is critical for ChIP success.<span> </span><br /></span></strong><strong><a href="../categories/chromatin-shearing"><span style="font-weight: 400;">Read more</span></a><span style="font-weight: 400;"><span> </span>about solutions for successful chromatin preparation.<span> </span><br /></span></strong><strong><span style="font-weight: 400;"><a href="../categories/bioruptor-shearing-device">Read more</a><span> </span>about<span> </span></span><span style="font-weight: 400;">Bioruptor<span> </span></span><span style="font-weight: 400;">sonication.</span></strong></p>
<p style="padding-left: 30px;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/immunoprecipitation.png" width="35" height="35" caption="false" /> Chromatin immunoprecipitation<br /></b></span></strong><span style="font-weight: 400;">Immunoprecipitation solutions for histone and transcription factor ChIP-qPCR and ChIP-seq for low inputs, plants, and animals including automated solutions</span><strong><span style="font-weight: 400;"><b><span style="font-weight: 400;">. <a href="../categories/chromatin-immunoprecipitation">Read more</a></span></b><br /></span></strong></p>
<p style="padding-left: 30px;"><strong><img src="https://www.diagenode.com/img/applications/ChIPmentation.png" alt="" width="35" height="35" /> ChIPmentation<br /><span style="font-weight: 400;">ChIPmentation, an exclusive technology, is an end-to-end ChIP-seq solution for low and difficult inputs. <a href="../categories/chromatin-ip-chipmentation">Read more</a></span><br /></strong></p>
<p style="padding-left: 30px;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/ChIP-antibodies.png" alt="" width="35" height="35" /> ChIP and ChIP-seq antibodies<br /></b></span></strong><span style="font-weight: 400;">ChIP-grade antibodies are essential for success. <a href="../categories/antibodies">Learn more</a></span><span style="font-weight: 400;"><br /></span></p>
<p style="padding-left: 30px;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/ChIP-qPCR.png" width="35" height="35" caption="false" /> Primer pairs<br /></b></span></strong><span style="font-weight: 400;">Highly specific primer pairs for the amplification of the specific genomic regions. <a href="../categories/primer-pairs">Read more</a></span><span style="font-weight: 400;"><br /></span></p>
<p style="padding-left: 30px;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/cutandtag.png" width="35" height="35" caption="false" /> CUT&Tag solutions<br /></b></span></strong><span style="font-weight: 400;">An alternative to ChIP-seq that combines antibody-targeted controlled cleavage by a protein A-Tn5 fusion with NGS to identify the binding sites of DNA-associated proteins. <a href="../categories/cutandtag">Read more</a></span><span style="font-weight: 400;"><br /></span></p>
<p style="padding-left: 30px;"><span style="font-weight: 400;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/dna-purification.png" width="35" height="35" caption="false" /> DNA purification<br /></b></span></strong><span style="font-weight: 400;"><a href="../categories/dna-and-rna-purification">Read more</a> about solutions for DNA purification</span></span></p>
<p style="padding-left: 30px;"><span style="font-weight: 400;"><span style="font-weight: 400;"><strong><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/ChIP-seq.png" width="35" height="35" caption="false" /> Library preparation for ChIP-seq<br /></b></span></strong><span style="font-weight: 400;">Optimized solutions for the library preparation from low DNA input. <a href="../categories/library-preparation-for-ChIP-seq">Read more</a></span></span></span></p>
<p style="padding-left: 30px;"><span style="font-weight: 400;"><b><img src="https://www.diagenode.com/img/applications/ChIP-automation.png" alt="" width="35" height="35" /> ChIP and ChIP-seq automation<br /></b>Reproducibility, optimization simplicity, no variability. <a href="../categories/epigenetic-automation">Learn more</a></span></p>
<div class="row" style="margin-top: 32px;">
<div class="small-12 medium-10 large-9 small-centered columns">
<div class="radius panel" style="background-color: #fff;">
<p class="text-justify">We offer <a href="https://www.diagenode.com/en/categories/chromatin-immunoprecipitation" target="_blank">complete ChIP kits</a> or <strong>individual kit components</strong> from antibodies, buffers, beads, chromatin shearing, and purification reagents. With the ChIP Kit Customizer, you have complete flexibility on which components you want from our validated ChIP kits.</p>
<div class="center" style="text-align: center;"><a href="../pages/chip-kit-customizer-1"><img src="https://www.diagenode.com/img/banners/banner-customizer.png" alt="" /></a></div>
</div>
</div>
</div>
<h4>Chromatin resources</h4>
<h3>Posters</h3>
<ul>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/antibody-chipseq-qc-using-the-ipstar-compact-poster"><span style="font-weight: 400;">Understanding our antibody QC</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/antibodies-you-can-trust-poster"><span style="font-weight: 400;">High quality ChIP antibodies</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/chip-kit-results-with-true-microchip-kit-poster"><span style="font-weight: 400;">ChIP with only 10,000 cells</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/high-resolution-chipseq-profiles-with-ipstar-automated-platform-poster"><span style="font-weight: 400;">High resolution ChIP-seq using automation</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/bioinformatics-pipeline-for-chipseq-analyses"><span style="font-weight: 400;">ChIP-seq bioinformatics</span></a></li>
</ul>
<h3><span style="font-weight: 400;">Application notes</span></h3>
<ul>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/chipettor-application-note"><span style="font-weight: 400;">Simple semi-automaton for easy and inexpensive ChIP</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/chipseq-from-human-tumor-tissue"><span style="font-weight: 400;">Performing ChIP-seq on human tumor tissue</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/plant-chip-seq-application-note"><span style="font-weight: 400;">Plant ChIP-seq – a successful method</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/documents/chip-seq-application-note"><span style="font-weight: 400;">Best workflow practices for low input ChIP</span></a></li>
<li style="font-weight: 400;"><a href="https://www.diagenode.com/files/application_notes/AN-ChIP-Cas9-02_2018.pdf"><span style="font-weight: 400;">Optimize the selection of guide RNA by ChIP to keep CRISPR on-target</span></a></li>
</ul>
<h3>Publications related to ChIP</h3>
<ul>
<li><a href="https://www.diagenode.com/en/publications/view/3373">Corticosteroid receptors adopt distinct cyclical transcriptional signatures</a></li>
<li><a href="https://www.diagenode.com/en/publications/view/3347">Pro-inflammatory cytokine and high doses of ionizing radiation have similar effects on the expression of NF-kappaB-dependent genes</a></li>
<li><a href="https://www.diagenode.com/en/publications/view/3355">Functional dissection of Drosophila melanogaster SUUR protein influence on H3K27me3 profile</a></li>
</ul>
<h3><span style="font-weight: 400;">Brochures</span></h3>
<ul>
<li><a href="https://www.diagenode.com/files/brochures/Chromatin_Immunoprecipitation_Brochure.pdf"><b>Chromatin </b><span style="font-weight: 400;">products brochure</span></a></li>
<li><a href="https://www.diagenode.com/files/brochures/Epigenetic_Antibodies_Brochure.pdf"><span style="font-weight: 400;">Epigenetic<span> </span></span><b>Antibodies</b></a></li>
<li><a href="https://www.diagenode.com/files/brochures/Bioruptor_Sonicator_Brochure.pdf"><span style="font-weight: 400;">Bioruptor for<span> </span></span><b>chromatin shearing</b></a></li>
<li><a href="https://www.diagenode.com/files/brochures/IPStar_Automated_System_Brochure.pdf"><span style="font-weight: 400;">Automating<span> </span></span><b>ChIP</b><span style="font-weight: 400;"><span> </span>and<span> </span></span><b>ChIP-seq</b></a></li>
</ul>
</div>
</div>',
'no_promo' => false,
'in_menu' => true,
'online' => true,
'tabular' => true,
'hide' => false,
'all_format' => false,
'is_antibody' => false,
'slug' => 'chromatin-function',
'cookies_tag_id' => null,
'meta_keywords' => 'Chromatin function,Chromatin shearing,Chromatin immunoprecipitation,Chromatin assembly',
'meta_description' => 'Diagenode offers a number of unique solutions and kits to make Diagenode offers chromatin immunoprecipitation kits have been designed to meet a wide variety of research needs and accessible',
'meta_title' => 'Chromatin Function Kit for Unique Solutions and Kits to make Chromatin Research| Diagenode',
'modified' => '2025-05-21 12:06:52',
'created' => '2015-05-27 05:22:15',
'ProductsCategory' => array(
[maximum depth reached]
),
'CookiesTag' => array([maximum depth reached])
)
),
'Document' => array(
(int) 0 => array(
'id' => '1139',
'name' => 'ATAC-seq Kit - Manual',
'description' => '<p class="p1">Gene expression is carefully regulated in the cells in order to manage a wide range of biological functions. The structure of chromatin is quite dynamic and contributes to this crucial regulatory process.</p>
<p class="p1">ATAC-seq, Assay for Transposase-Accessible Chromatin, followed by nextgeneration sequencing, is a key technology to easily identify the “open” regions of the chromatin, which are usually associated with permissive gene expression. Indeed, the nuclei of the samples are incubated with a transposase, and only the genomic regions associated with open chromatin will be accessible to this transposase. During the process those regions will be cut and sequencing adaptors will be added, allowing their sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only., giving a map of the chromatin status in the whole genome of the sample.</p>
<p class="p1">The Diagenode’s ATAC-seq kit is based on a highly validated protocol, used for years in our Epigenomics Profiling Services offer and takes advantage of many successful Diagenode’s tools, such as the loaded Tagmentase (Tn5 transposase), the MicroChIP DiaPure Columns and the Primer indexes for tagmented libraries kits.</p>',
'image_id' => null,
'type' => 'Manual',
'url' => 'files/products/kits/atacseq-kit-manual.pdf',
'slug' => 'atacseq-kit-manual',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-06-25 09:51:59',
'created' => '2021-06-25 09:51:59',
'ProductsDocument' => array(
[maximum depth reached]
)
)
),
'Feature' => array(),
'Image' => array(
(int) 0 => array(
'id' => '1819',
'name' => 'product/kits/atacseq-kit-picture.jpg',
'alt' => 'ATAC-seq kit',
'modified' => '2021-06-25 10:03:22',
'created' => '2021-06-25 10:03:22',
'ProductsImage' => array(
[maximum depth reached]
)
)
),
'Promotion' => array(),
'Protocol' => array(),
'Publication' => array(
(int) 0 => array(
'id' => '5143',
'name' => 'CRAMP1 drives linker histone expression to enable Polycomb repression',
'authors' => 'Matthews, Rachael E et al.',
'description' => '<div class="abstract-content selected" id="eng-abstract">
<p style="text-align: justify;">In contrast to the well-understood role of core histones in DNA packaging, the function of the linker histone (H1) remains enigmatic. Challenging the prevailing view that linker histones are a general feature of heterochromatin, here we show a critical requirement for H1 in Polycomb repressive complex 2 (PRC2) function. A CRISPR-Cas9 genetic screen using a fluorescent PRC2 reporter identified an essential role for the poorly characterized gene CRAMP1 in PRC2-mediated repression. CRAMP1 localizes to the promoters of expressed H1 genes and positively regulates their transcription. CRAMP1 ablation simultaneously depletes all linker histones, which results in selective decompaction of H3K27me3-marked loci and derepression of PRC2 target genes without concomitant loss of PRC2 occupancy or enzymatic activity. Strikingly, we find that linker histones preferentially localize to genomic loci marked by H3K27me3 across diverse cell types and organisms. Altogether, these data demonstrate a prominent role for linker histones in epigenetic repression by PRC2.</p>
</div>',
'date' => '2025-06-10',
'pmid' => 'https://pubmed.ncbi.nlm.nih.gov/40516528/',
'doi' => '10.1016/j.molcel.2025.05.031',
'modified' => '2025-06-18 09:45:01',
'created' => '2025-06-18 09:45:01',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '5142',
'name' => 'Temporal and spatial niche partitioning in a retrotransposon community of the Drosophila melanogaster genome',
'authors' => 'Varoqui, Marion et al.',
'description' => '<div class="abstract" id="abstract">
<div class="abstract-content selected" id="eng-abstract">
<p style="text-align: justify;">Transposable elements (TEs) are genetic parasites that can potentially threaten the stability of the genomes they colonize. Nonetheless, TEs persist within genomes and are rarely fully eliminated, with diverse TE families coexisting in varing copy numbers. The TE replication strategies that enable host organisms to tolerate and accommodate the extensive diversity of TEs, while minimizing harm to the host and avoiding mutual competition among TEs, remain poorly understood. Here, by studying the spontaneous or experimental mobilization of four Drosophila LTR RetroTransposable Elements (LTR-RTEs), we reveal that each of them preferentially targets open chromatin regions characterized by specific epigenetic features. Among these, gtwin and ZAM are expressed in distinct cell types within female somatic gonadal tissues and inserted into the distinct accessible chromatin landscapes of the corresponding stages of embryogenesis. These findings suggest that individual LTR-RTEs exploit unique biological niches, enabling their coexistence within the tightly regulated ecosystem of the same host genome.</p>
</div>
</div>',
'date' => '2025-06-06',
'pmid' => 'https://pubmed.ncbi.nlm.nih.gov/40503684/',
'doi' => '10.1093/nar/gkaf516',
'modified' => '2025-06-18 09:43:08',
'created' => '2025-06-18 09:43:08',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '5128',
'name' => 'Cervicovaginal microbiome alters transcriptomic and epigenomic signatures across cervicovaginal epithelial barriers',
'authors' => 'Lauren Anton et al. ',
'description' => '<p><b>Background</b></p>
<p>The cervicovaginal microbiome plays a critical role in women's health, with microbial communities dominated by<span> </span><em>Lactobacillus</em><span> </span>species considered optimal. In contrast, the depletion of lactobacilli and the presence of a diverse array of strict and facultative anaerobes, such as<span> </span><em>Gardnerella vaginalis</em>, have been linked with adverse reproductive outcomes. Despite these associations, the molecular mechanisms by which host-microbial interactions modulate cervical and vaginal epithelial function remains poorly understood.</p>
<p><b>Results</b></p>
<p>In this study, we used RNA sequencing to characterize the transcriptional response of cervicovaginal epithelial cells exposed to the culture supernatants of common vaginal bacteria. Our findings revealed that<span> </span><em>G. vaginalis</em><span> </span>culture supernatants upregulate genes associated with an activated innate immune response and increased cell death. Conversely,<span> </span><em>Lactobacillus crispatus</em><span> </span>culture supernatants induced transcriptional changes indicative of epigenomic modeling in ectocervical epithelial cells. Epigenomic modification by<span> </span><em>L. crispatus</em>, was confirmed by ATAC-sequencing, which demonstrated reduced chromatin accessibility.</p>
<p><b>Conclusions</b></p>
<p>These results provide new insights into host-microbe interactions within the lower reproductive tract and suggests that modulating the vaginal microbiome could offer innovative therapeutic strategies to improve reproductive health.</p>',
'date' => '2025-05-07',
'pmid' => 'https://www.researchsquare.com/article/rs-6171614/v1',
'doi' => 'https://doi.org/10.21203/rs.3.rs-6171614/v1',
'modified' => '2025-05-12 10:30:24',
'created' => '2025-05-12 10:30:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '5125',
'name' => 'An ISWI-related chromatin remodeller regulates stage-specific gene expression in Toxoplasma gondii',
'authors' => 'Pachano, Belen et al.',
'description' => '<p style="text-align: justify;"><span>ATP-dependent chromatin remodellers are specialized multiprotein machines that organize the genome in eukaryotic cells and regulate its accessibility by repositioning, ejecting or modifying nucleosomes. However, their role in </span><i>Toxoplasma gondii</i><span><span> </span>is poorly understood. Here we show that<span> </span></span><i>T.</i><span> </span><i>gondii</i><span><span> </span>has evolved two divergent proteins within the imitation switch (ISWI) family:<span> </span></span><i>Tg</i><span>SNF2h and<span> </span></span><i>Tg</i><span>SNF2L.<span> </span></span><i>Tg</i><span>SNF2h specifically forms a core complex with the transcription factor AP2VIII-2 and the scaffold protein<span> </span></span><i>Tg</i><span>RFTS. Depletion of<span> </span></span><i>Tg</i><span>RFTS phenocopies the knockdown of<span> </span></span><i>Tg</i><span>SNF2h, restricting access to chromatin and altering local gene expression. At the genomic level,<span> </span></span><i>Tg</i><span>SNF2h insulates highly transcribed genes from silenced neighbours, ensuring stage-specific gene regulation. By modulating chromatin accessibility to transcription factors,<span> </span></span><i>Tg</i><span>SNF2h exerts epistatic control over MORC, a key regulator of sexual commitment. Our findings show that a specific ISWI complex orchestrates the partitioning of developmental genes and ensures transcriptional fidelity throughout the parasite life cycle.</span></p>',
'date' => '2025-04-11',
'pmid' => 'https://www.nature.com/articles/s41564-025-01980-2#citeas',
'doi' => 'https://doi.org/10.1038/s41564-025-01980-2',
'modified' => '2025-05-19 11:36:47',
'created' => '2025-05-06 15:49:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '5101',
'name' => 'Repression of oxidative phosphorylation by NR2F2, MTERF3 and GDF15 in human skin under high-glucose stress',
'authors' => 'S. Ley-Ngardigal et al.',
'description' => '<p><span>Lifestyle factors such as a Western diet or metabolic diseases like diabetes disrupt glucose homeostasis and induce stress responses, yet their impact on skin metabolism and structural integrity remains poorly understood. Here, we performed multiomic and bioenergetic analyses of human dermal fibroblasts (HDFs), human equivalent dermis (HED), human reconstructed skin (HRS), and skin explants from diabetic patients. We found that 12 mM glucose stress represses oxidative phosphorylation (OXPHOS) through a dual mechanism: the glucose-dependent nuclear receptor NR2F2 activates mitochondrial transcription termination factor 3 (MTERF3) while inhibiting growth-differentiation factor 15 (GDF15). Promoter assays revealed that MTERF3 is regulated by NR2F2 and MYCN, whereas GDF15 is modulated by NR2F2 and FOS. Consequently, OXPHOS proteins and mitochondrial respiration were suppressed, and MTERF3 overexpression additionally interfered with collagen biosynthesis. In contrast, GDF15 supplementation fully rescued hyperglycemia-induced bioenergetic and metabolomic alterations, suggesting a pharmacological strategy to mitigate hyperglycemic damage in the skin. Finally, silencing GDF15 or TFAM impaired fibroblast haptotaxis and skin reconstruction, underscoring the crucial role of mitochondrial energetics in dermal structure and function. Collectively, these findings identify the NR2F2–MTERF3–GDF15 axis as a key mediator of OXPHOS suppression and highlight a potential therapeutic target to preserve skin integrity under hyperglycemic stress.</span></p>',
'date' => '2025-03-27',
'pmid' => 'https://www.sciencedirect.com/science/article/pii/S2213231725001260',
'doi' => 'https://doi.org/10.1016/j.redox.2025.103613',
'modified' => '2025-03-31 13:44:10',
'created' => '2025-03-31 13:44:10',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '5092',
'name' => 'CRISPR screening identifies regulators of enhancer-mediated androgen receptor transcription in advanced prostate cancer',
'authors' => 'Rachel R. Xiang et al. ',
'description' => '<section id="author-highlights-abstract" property="abstract" typeof="Text" role="doc-abstract">
<h2 property="name"></h2>
</section>
<section id="author-abstract" property="abstract" typeof="Text" role="doc-abstract">
<div id="abspara0010" role="paragraph">Amplification of the androgen receptor (AR) locus is the most frequent alteration in metastatic castration-resistant prostate cancer (CRPC). Recently, it was discovered that an enhancer of the AR is co-amplified with the AR gene body and contributes to increased AR transcription and resistance to androgen deprivation therapy. However, the mechanism of enhancer activation in advanced disease is unknown. Here, we used CRISPR-Cas9 screening to identify transcription factors that bind to the AR enhancer and modulate enhancer-mediated AR transcription. We demonstrate that HOXB13, GATA2, and TFAP2C bind the AR enhancer in patient-derived xenografts and directly impact features associated with an active chromatin state. Interestingly, the AR enhancer belongs to a set of regulatory elements that require HOXB13 to maintain FOXA1 binding, further delineating the role of HOXB13 in CRPC. This work provides a framework to functionally identify<span> </span><i>trans</i>-acting factors required for the activation of disease-related noncoding regulatory elements.</div>
</section>',
'date' => '2025-02-14',
'pmid' => 'https://www.cell.com/cell-reports/fulltext/S2211-1247(25)00083-X',
'doi' => '10.1016/j.celrep.2025.115312',
'modified' => '2025-03-27 16:56:52',
'created' => '2025-03-27 16:54:22',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '5038',
'name' => 'Schistosoma mansoni antigen induced innate immune memory features mitochondrial biogenesis and can be inhibited by ovarian produced hormones',
'authors' => 'Juan Marcos Oviedo et al.',
'description' => '<p><span>We have previously identified that </span><em>S. mansoni</em><span><span> </span>infection induces a unique form of myeloid training that protects male but not female mice from high fat diet induced disease. Here we demonstrate that ovarian derived hormones account for this sex specific difference. Ovariectomy of females prior to infection permits metabolic reprogramming of the myeloid lineage, with BMDM exhibiting carbon source flexibility for cellular respiration, and mice protected from systemic metabolic disease. The innate training phenotype of infection can be replicated by<span> </span></span><em>in vivo</em><span><span> </span>injection of SEA, and by exposure of bone marrow to SEA in culture prior to macrophage differentiation (Day 0). This protective phenotype is linked to increased chromatin accessibility of lipid and mitochondrial pathways in BMDM including Nrf1 and Tfam, as well as mitochondrial biogenesis. This work provides evidence that<span> </span></span><em>S. mansoni</em><span><span> </span>antigens induce a unique form of innate training inhibited by ovarian-derived hormones in females.</span></p>',
'date' => '2025-01-18',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2025.01.14.632838v1',
'doi' => 'https://doi.org/10.1101/2025.01.14.632838',
'modified' => '2025-01-31 10:55:42',
'created' => '2025-01-31 10:55:42',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '5117',
'name' => 'Dynamic PRC1–CBX8 stabilizes a porous structure of chromatin condensates',
'authors' => 'Uckelmann, M., et al.',
'description' => '<p><strong>Abstract</strong></p>
<div class="c-article-section__content" id="Abs1-content">
<p style="text-align: justify;">The compaction of chromatin is a prevalent paradigm in gene repression. Chromatin compaction is commonly thought to repress transcription by restricting chromatin accessibility. However, the spatial organization and dynamics of chromatin compacted by gene-repressing factors are unknown. Here, using cryo-electron tomography, we solved the three-dimensional structure of chromatin condensed by the polycomb repressive complex 1 (PRC1) in a complex with CBX8. PRC1-condensed chromatin is porous and stabilized through multivalent dynamic interactions of PRC1 with chromatin. Mechanistically, positively charged residues on the internally disordered regions of CBX8 mask negative charges on the DNA to stabilize the condensed state of chromatin. Within condensates, PRC1 remains dynamic while maintaining a static chromatin structure. In differentiated mouse embryonic stem cells, CBX8-bound chromatin remains accessible. These findings challenge the idea of rigidly compacted polycomb domains and instead provide a mechanistic framework for dynamic and accessible PRC1–chromatin condensates.</p>
</div>',
'date' => '2025-01-15',
'pmid' => 'https://www.nature.com/articles/s41594-024-01457-6',
'doi' => 'https://doi.org/10.1038/s41594-024-01457-6',
'modified' => '2025-04-25 13:20:40',
'created' => '2025-04-25 11:48:09',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 8 => array(
'id' => '5076',
'name' => 'Assessing the impact of whole genome duplication on gene expression and regulation during arachnid development',
'authors' => 'Madeleine E. Aase-Remedios et al.',
'description' => '<p id="p-2">Whole genome duplication (WGD) generates new genetic material that can contribute to the evolution of the regulation of developmental processes and phenotypic diversification. A WGD occurred in an ancestor of arachnopulmonates (spiders, scorpions, and their relatives), which provides an important independent comparison to WGDs in other animal lineages like those in vertebrates and horseshoe crabs. After WGD, arachnopulmonates retained many duplicated copies (ohnologues) of developmental genes including clusters of homeobox genes, many of which have been inferred to have undergone subfunctionalisation. However, there has been little systematic analysis of gene regulatory sequences and comparison of the expression of ohnologues versus their single-copy orthologues between arachnids. Here we compare the regions of accessible chromatin and gene expression of ohnologues and single-copy genes during three embryonic stages between an arachnopulmonate arachnid, the spider<span> </span><em>Parasteatoda tepidariorum</em>, and a non-arachnopulmonate arachnid, the harvestman<span> </span><em>Phalangium opilio</em>. We found that the expression of each spider ohnologue was lower than their single-copy orthologues in the harvestman providing evidence for subfunctionalisation. However, this was not reflected in a reduction in peaks of accessible chromatin because both spider ohnologues and single-copy genes had more peaks than the harvestman genes. We also found peaks of open chromatin that increased in the late stage associated with activation of genes expressed later during embryogenesis in both species. Taken together, our study provides the first genome-wide comparison of gene regulatory sequences and gene expression in arachnids and thus provides new insights into the impact of the arachnopulmonate WGD.</p>
<div id="sec-1" class="subsection">
<p id="p-3"><strong>Significance statement</strong><span> </span>The comparison of independent WGD events is essential to understanding their evolutionary outcomes. Here we examine gene expression and chromatin accessibility during the development of two arachnids, one of which underwent an ancient WGD. Our data provide the first direct comparison of the regulation of ohnologues and single-copy genes in arachnids.</p>
</div>',
'date' => '2024-12-20',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2024.12.18.628675v1.full',
'doi' => 'https://doi.org/10.1101/2024.12.18.628675',
'modified' => '2025-02-27 17:24:20',
'created' => '2025-02-27 17:24:20',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 9 => array(
'id' => '5012',
'name' => 'Spatial organizations of heterochromatin underpin nuclear structural integrity of ventricular cardiomyocytes against mechanical stress',
'authors' => 'Keita Fujiwara et al.',
'description' => '<section id="author-highlights-abstract" property="abstract" typeof="Text" role="doc-abstract">
<h2 property="name">Highlights</h2>
<div id="abspara0020" role="paragraph">
<div id="ulist0010" role="list">
<div id="u0010" role="listitem">
<div class="content">
<div id="p0010" role="paragraph">Cardiomyocytes acquire characteristic spatial organizations of heterochromatin (SOH)</div>
</div>
</div>
<div id="u0015" role="listitem">
<div class="content">
<div id="p0015" role="paragraph">High levels of H2B-mCherry disrupt SOH, leading to nuclear elongation in cardiomyocytes</div>
</div>
</div>
<div id="u0020" role="listitem">
<div class="content">
<div id="p0020" role="paragraph">SOH disruption minimally impacts gene expression despite loosening global genome structure</div>
</div>
</div>
<div id="u0025" role="listitem">
<div class="content">
<div id="p0025" role="paragraph">SOH alter with aging, leading to nuclear deformation in ventricular cardiomyocytes</div>
</div>
</div>
</div>
</div>
</section>
<section id="author-abstract" property="abstract" typeof="Text" role="doc-abstract">
<h2 property="name">Summary</h2>
<div id="abspara0010" role="paragraph">Cardiomyocyte (CM) nuclei are constantly exposed to mechanical stress, but how they maintain their nuclear shape remains unknown. In this study, we found that ventricular CM nuclei acquire characteristic prominent spatial organizations of heterochromatin (SOH), which are disrupted by high-level expression of H2B-mCherry in mice. SOH disruption was associated with nuclear softening, leading to extreme elongation and rupture under unidirectional mechanical stress. Loosened chromatin then leaks into the cytosol, causing severe inflammation and cardiac dysfunction. Although SOH disruption was accompanied by loosened higher-order genomic structures, the change in gene expression before nuclear deformation was mild, suggesting that SOH play major roles in nuclear structural integrity. Aged CM nuclei consistently exhibited scattered SOH and marked elongation. Furthermore, we provide mechanistic insight into the development and maintenance of SOH driven by chromatin compaction and condensate formation. These results highlight SOH as a safeguard of nuclear shape and genomic integrity against mechanical stress.</div>
</section>',
'date' => '2024-12-09',
'pmid' => 'https://www.cell.com/cell-reports/fulltext/S2211-1247(24)01399-8',
'doi' => '10.1016/j.celrep.2024.115048',
'modified' => '2024-12-12 15:04:47',
'created' => '2024-12-12 15:04:47',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 10 => array(
'id' => '4984',
'name' => 'DNA demethylation triggers cell free DNA release in colorectal cancer cells',
'authors' => 'Valeria Pessei et al.',
'description' => '<h3 class="c-article__sub-heading" data-test="abstract-sub-heading">Background</h3>
<p>Liquid biopsy based on cell-free DNA (cfDNA) analysis holds significant promise as a minimally invasive approach for the diagnosis, genotyping, and monitoring of solid malignancies. Human tumors release cfDNA in the bloodstream through a combination of events, including cell death, active and passive release. However, the precise mechanisms leading to cfDNA shedding remain to be characterized. Addressing this question in patients is confounded by several factors, such as tumor burden extent, anatomical and vasculature barriers, and release of nucleic acids from normal cells. In this work, we exploited cancer models to dissect basic mechanisms of DNA release.</p>
<h3 class="c-article__sub-heading" data-test="abstract-sub-heading">Methods</h3>
<p>We measured cell loss ratio, doubling time, and cfDNA release in the supernatant of a colorectal cancer (CRC) cell line collection (<i>N</i> = 76) representative of the molecular subtypes previously identified in cancer patients. Association analyses between quantitative parameters of cfDNA release, cell proliferation, and molecular features were evaluated. Functional experiments were performed to test the impact of modulating DNA methylation on cfDNA release.</p>
<h3 class="c-article__sub-heading" data-test="abstract-sub-heading">Results</h3>
<p>Higher levels of supernatant cfDNA were significantly associated with slower cell cycling and increased cell death. In addition, a higher cfDNA shedding was found in non-CpG Island Methylator Phenotype (CIMP) models. These results indicate a positive correlation between lower methylation and increased cfDNA levels. To explore this further, we exploited methylation microarrays to identify a subset of probes significantly associated with cfDNA shedding and derive a methylation signature capable of discriminating high from low cfDNA releasers. We applied this signature to an independent set of 176 CRC cell lines and patient derived organoids to select 14 models predicted to be low or high releasers. The methylation profile successfully predicted the amount of cfDNA released in the supernatant. At the functional level, genetic ablation of DNA methyl-transferases increased chromatin accessibility and DNA fragmentation, leading to increased cfDNA release in isogenic CRC cell lines. Furthermore, in vitro treatment of five low releaser CRC cells with a demethylating agent was able to induce a significant increase in cfDNA shedding.</p>
<h3 class="c-article__sub-heading" data-test="abstract-sub-heading">Conclusions</h3>
<p>Methylation status of cancer cell lines contributes to the variability of cfDNA shedding in vitro. Changes in methylation pattern are associated with cfDNA release levels and might be exploited to increase sensitivity of liquid biopsy assays.</p>',
'date' => '2024-10-09',
'pmid' => 'https://link.springer.com/article/10.1186/s13073-024-01386-5',
'doi' => 'https://doi.org/10.1186/s13073-024-01386-5',
'modified' => '2024-10-14 08:56:24',
'created' => '2024-10-14 08:56:24',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 11 => array(
'id' => '4967',
'name' => 'Temporal and spatial niche partitioning in a retrotransposon community of the Drosophila genome',
'authors' => 'Varoqui M. et al.',
'description' => '<p><span>Transposable elements (TEs), widespread genetic parasites, pose potential threats to the stability of their host genomes. Hence, the interactions observed today between TEs and their host genomes, as well as among the different TE species coexisting in the same host, likely reflect those that did not lead to the extinction of either the host or the TEs. It is not clear to what extent the expression and integration steps of the TE replication cycles are involved in this ‘peaceful’ coexistence. Here, we show that four Drosophila LTR RetroTransposable Elements (LTR-RTEs), although sharing the same overall integration mechanism, preferentially integrate into distinct open chromatin domains of the host germline. Notably, the differential expressions of the gtwin and ZAM LTR-RTEs in ovarian and embryonic somatic tissues, respectively, result in differential integration timings and targeting of accessible chromatin landscapes that differ between early and late embryonic nuclei, highlighting connections between temporal and spatial LTR-RTEs niche partitionings.</span></p>',
'date' => '2024-08-16',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2024.08.14.607943v1',
'doi' => 'https://doi.org/10.1101/2024.08.14.607943',
'modified' => '2024-09-02 10:29:44',
'created' => '2024-09-02 10:29:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 12 => array(
'id' => '5093',
'name' => 'Galectin 3-binding protein (LGALS3BP) depletion attenuates hepatic fibrosis by reducing transforming growth factor-β1 (TGF-β1) availability and inhibits hepatocarcinogenesis',
'authors' => 'Dae-Hwan Kim et al.',
'description' => '<section class="article-section__content" id="cac212600-sec-0010">
<h3 class="article-section__sub-title section1" id="cac212600-sec-0010-title">Background</h3>
<p>Increased Galectin 3-binding protein (LGALS3BP) serum levels have been used to assess hepatic fibrosis stages and the severity of hepatocellular carcinoma (HCC). Considering the crucial role of transforming growth factor-β1 (TGF-β1) in the emergence of these diseases, the present study tested the hypothesis that LGALS3BP regulates the TGF-β1 signaling pathway.</p>
</section>
<section class="article-section__content" id="cac212600-sec-0020">
<h3 class="article-section__sub-title section1" id="cac212600-sec-0020-title">Methods</h3>
<p>The expression levels of<span> </span><i>LGALS3BP</i><span> </span>and<span> </span><i>TGFB1</i><span> </span>were analyzed in patients with metabolic dysfunction-associated steatohepatitis (MASH) and HCC. Multiple omics techniques, such as RNA-sequencing, transposase-accessible chromatin-sequencing assay, and liquid chromatography-tandem mass spectrometry proteomics, were used to identify the regulatory mechanisms for the LGALS3BP-TGF-β1 axis. The effects of altered TGF-β1 signaling by LGALS3BP were investigated in conditional<span> </span><i>LGALS3BP</i>-knockin and<span> </span><i>LGALS3BP</i>-knockout mice.</p>
</section>
<section class="article-section__content" id="cac212600-sec-0030">
<h3 class="article-section__sub-title section1" id="cac212600-sec-0030-title">Results</h3>
<p>In patients with MASH and HCC, the levels of<span> </span><i>LGALS3BP</i><span> </span>and<span> </span><i>TGFB1</i><span> </span>exhibited positive correlations. Stimulation of LGALS3BP by the inflammatory cytokine interferon α in HCC cells or ectopic overexpression of LGALS3BP in hepatocytes promoted the expression levels of TGFB1. Aggravated fibrosis was observed in the livers of hepatocyte-specific<span> </span><i>LGALS3BP</i>-knockin mice, with increased TGFB1 levels. LGALS3BP directly bound to and assembled integrin αV, an integral mediator required for releasing active TGF-β1 from extracellular latent complex with the rearranged F-actin cytoskeleton. The released TGF-β1 activated JunB transcription factor, which in turn promoted the TGF-β1 positive feedback loop.<span> </span><i>LGALS3BP</i><span> </span>deletion in the hepatocytes downregulated TGF-β1 signaling and CCl<sub>4</sub><span> </span>induced fibrosis. Moreover,<span> </span><i>LGALS3BP</i><span> </span>depletion hindered hepatocarcinogenesis by limiting the availability of fibrogenic TGF-β1.</p>
</section>
<section class="article-section__content" id="cac212600-sec-0040">
<h3 class="article-section__sub-title section1" id="cac212600-sec-0040-title">Conclusion</h3>
<p>LGALS3BP plays a crucial role in hepatic fibrosis and carcinogenesis by controlling the TGF-β1 signaling pathway, making it a promising therapeutic target in TGF-β1-related diseases.</p>
</section>',
'date' => '2024-07-28',
'pmid' => 'https://onlinelibrary.wiley.com/doi/full/10.1002/cac2.12600',
'doi' => ' https://doi.org/10.1002/cac2.12600',
'modified' => '2025-03-27 16:59:26',
'created' => '2025-03-27 16:59:26',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 13 => array(
'id' => '5091',
'name' => 'Patho-transcriptomic analysis of invasive mucinous adenocarcinoma of the lung (IMA): comparison with lung adenocarcinoma with signet ring cell features (SRCC)',
'authors' => 'William D. Stuart et al.',
'description' => '<div id="sec-1" class="subsection">
<p id="p-3"><strong>Background</strong><span> </span>Invasive mucinous adenocarcinoma (IMA) comprises ∼5% of lung adenocarcinoma. There is no effective therapy for IMA when surgical resection is not possible. IMA is sometimes confused with adenocarcinoma with signet ring cell features (SRCC) pathologically since both adenocarcinomas feature tumor cells with abundant intracellular mucin. The molecular mechanisms by which such mucin-producing lung adenocarcinomas develop remain unknown.</p>
</div>
<div id="sec-2" class="subsection">
<p id="p-4"><strong>Methods</strong><span> </span>Using a Visium spatial transcriptomics approach, we analyzed IMA and compared it with SRCC patho-transcriptomically. Combining spatial transcriptomics data with<span> </span><em>in vitro</em><span> </span>studies using RNA-seq and ChIP-seq, we assessed downstream targets of transcription factors HNF4A and SPDEF that are highly expressed in IMA and/or SRCC.</p>
</div>
<div id="sec-3" class="subsection">
<p id="p-5"><strong>Results</strong><span> </span>Spatial transcriptomics analysis indicated that there are 6 distinct cell clusters in IMA and SRCC. Notably, two clusters (C1 and C3) of mucinous tumor cells exist in both adenocarcinomas albeit at a different ratio. Importantly, a portion of genes (e.g.,<span> </span><em>NKX2-1</em>,<span> </span><em>GKN1</em>,<span> </span><em>HNF4A</em><span> </span>and<span> </span><em>FOXA3</em>) are distinctly expressed while some mucous-related genes (e.g.,<span> </span><em>SPDEF</em><span> </span>and<span> </span><em>FOXA2</em>) are expressed in both adenocarcinomas. We determined that HNF4A induces<span> </span><em>MUC3A/B</em><span> </span>and<span> </span><em>TM4SF4</em><span> </span>and that BI 6015, an HNF4A antagonist, suppressed the growth of IMA cells. Using mutant SPDEF that is associated with COVID-19, we also determined that an intact DNA-binding domain of SPDEF is required for SPDEF-mediated induction of mucin genes (<em>MUC5AC</em>,<span> </span><em>MUC5B</em><span> </span>and<span> </span><em>AGR2</em>). Additionally, we found that XMU-MP-1, a SPDEF inhibitor, suppressed the growth of IMA cells.</p>
</div>
<div id="sec-4" class="subsection">
<p id="p-6"><strong>Conclusion</strong><span> </span>These results revealed that IMA and SRCC contain heterogenous tumor cell types, some of which are targetable.</p>
</div>',
'date' => '2024-06-17',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2024.06.13.598839v1.full',
'doi' => 'https://doi.org/10.1101/2024.06.13.598839',
'modified' => '2025-03-27 16:08:19',
'created' => '2025-03-27 16:08:19',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 14 => array(
'id' => '5094',
'name' => 'Transient loss of Polycomb components induces an epigenetic cancer fate',
'authors' => 'V. Parreno et al.',
'description' => '<p><span>Although cancer initiation and progression are generally associated with the accumulation of somatic mutations</span><sup><a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 1" title="McGranahan, N. & Swanton, C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 27, 15–26 (2015)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR1" id="ref-link-section-d121858e719">1</a>,<a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 2" title="Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR2" id="ref-link-section-d121858e722">2</a></sup><span>, substantial epigenomic alterations underlie many aspects of tumorigenesis and cancer susceptibility</span><sup><a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" title="Brock, A., Chang, H. & Huang, S. Non-genetic heterogeneity—a mutation-independent driving force for the somatic evolution of tumours. Nat. Rev. Genet. 10, 336–342 (2009)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR3" id="ref-link-section-d121858e726">3</a>,<a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" title="Flavahan, W. A., Gaskell, E. & Bernstein, B. E. Epigenetic plasticity and the hallmarks of cancer. Science 357, eaal2380 (2017)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR4" id="ref-link-section-d121858e726_1">4</a>,<a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" title="Marine, J. C., Dawson, S. J. & Dawson, M. A. Non-genetic mechanisms of therapeutic resistance in cancer. Nat. Rev. Cancer 20, 743–756 (2020)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR5" id="ref-link-section-d121858e726_2">5</a>,<a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 6" title="Timp, W. & Feinberg, A. P. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat. Rev. Cancer 13, 497–510 (2013)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR6" id="ref-link-section-d121858e729">6</a></sup><span>, suggesting that genetic mechanisms might not be the only drivers of malignant transformation</span><sup><a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 7" title="Teixeira, V. H. et al. Deciphering the genomic, epigenomic, and transcriptomic landscapes of pre-invasive lung cancer lesions. Nat. Med. 25, 517–525 (2019)." href="https://www.nature.com/articles/s41586-024-07328-w#ref-CR7" id="ref-link-section-d121858e733">7</a></sup><span>. However, whether purely non-genetic mechanisms are sufficient to initiate tumorigenesis irrespective of mutations has been unknown. Here, we show that a transient perturbation of transcriptional silencing mediated by Polycomb group proteins is sufficient to induce an irreversible switch to a cancer cell fate in<span> </span></span><i>Drosophila</i><span>. This is linked to the irreversible derepression of genes that can drive tumorigenesis, including members of the JAK–STAT signalling pathway and<span> </span></span><i>zfh1</i><span>, the fly homologue of the<span> </span></span><i>ZEB1</i><span><span> </span>oncogene, whose aberrant activation is required for Polycomb perturbation-induced tumorigenesis. These data show that a reversible depletion of Polycomb proteins can induce cancer in the absence of driver mutations, suggesting that tumours can emerge through epigenetic dysregulation leading to inheritance of altered cell fates.</span></p>',
'date' => '2024-04-24',
'pmid' => 'https://www.nature.com/articles/s41586-024-07328-w',
'doi' => 'https://doi.org/10.1038/s41586-024-07328-w',
'modified' => '2025-03-27 17:00:44',
'created' => '2025-03-27 17:00:44',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 15 => array(
'id' => '4933',
'name' => 'Systematic mapping of TF-mediated cell fate changes by a pooled induction coupled with scRNA-seq and multi-omics approaches',
'authors' => 'Lee M. et al.',
'description' => '<p><span>Transcriptional regulation controls cellular functions through interactions between transcription factors (TFs) and their chromosomal targets. However, understanding the fate conversion potential of multiple TFs in an inducible manner remains limited. Here, we introduce iTF-seq as a method for identifying individual TFs that can alter cell fate toward specific lineages at a single-cell level. iTF-seq enables time course monitoring of transcriptome changes, and with biotinylated individual TFs, it provides a multi-omics approach to understanding the mechanisms behind TF-mediated cell fate changes. Our iTF-seq study in mouse embryonic stem cells identified multiple TFs that trigger rapid transcriptome changes indicative of differentiation within a day of induction. Moreover, cells expressing these potent TFs often show a slower cell cycle and increased cell death. Further analysis using bioChIP-seq revealed that GCM1 and OTX2 act as pioneer factors and activators by increasing gene accessibility and activating the expression of lineage specification genes during cell fate conversion. iTF-seq has utility in both mapping cell fate conversion and understanding cell fate conversion mechanisms.</span></p>',
'date' => '2024-04-05',
'pmid' => 'https://pubmed.ncbi.nlm.nih.gov/38580401/',
'doi' => '10.1101/gr.277926.123',
'modified' => '2024-04-09 00:19:18',
'created' => '2024-04-09 00:19:18',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 16 => array(
'id' => '4895',
'name' => 'Protocol to isolate nuclei from Chlamydomonas reinhardtii for ATAC sequencing',
'authors' => 'Santhanagopalan I. et al.',
'description' => '<p class="section-title u-h4 u-margin-l-top u-margin-xs-bottom">Highlights</p>
<div id="abssec0020">
<ul class="list">
<li class="react-xocs-list-item">
<p id="p0010">Optimized isolation of nuclei from the green model alga<span> </span><em>Chlamydomonas reinhardtii</em></p>
</li>
<li class="react-xocs-list-item">
<p id="p0010"><em></em></p>
<p id="p0015">Tag-free isolation from both cell-walled and cell wall-deficient algae strains</p>
</li>
<li class="react-xocs-list-item">
<p id="p0015"></p>
<p id="p0020">Key steps for an effective and fast isolation and quantification procedure of nuclei</p>
</li>
<li class="react-xocs-list-item">
<p><span class="list-label"></span>Extracts at a quality suitable for ATAC-sequencing</p>
</li>
</ul>
</div>',
'date' => '2024-03-15',
'pmid' => 'https://www.sciencedirect.com/science/article/pii/S2666166723007311',
'doi' => 'https://doi.org/10.1016/j.xpro.2023.102764',
'modified' => '2024-02-09 12:37:32',
'created' => '2024-01-22 13:39:58',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 17 => array(
'id' => '4917',
'name' => 'Interplay between coding and non-coding regulation drives the Arabidopsis seed-to-seedling transition',
'authors' => 'Trembley B.J.M. et al.',
'description' => '<p><span>Translation of seed stored mRNAs is essential to trigger germination. However, when RNAPII re-engages RNA synthesis during the seed-to-seedling transition has remained in question. Combining csRNA-seq, ATAC-seq and smFISH in </span><i>Arabidopsis thaliana</i><span><span> </span>we demonstrate that active transcription initiation is detectable during the entire germination process. Features of non-coding regulation such as dynamic changes in chromatin accessible regions, antisense transcription, as well as bidirectional non-coding promoters are widespread throughout the Arabidopsis genome. We show that sensitivity to exogenous ABSCISIC ACID (ABA) during germination depends on proximal promoter accessibility at ABA-responsive genes. Moreover, we provide genetic validation of the existence of divergent transcription in plants. Our results reveal that active enhancer elements are transcribed producing non-coding enhancer RNAs (eRNAs) as widely documented in metazoans. In sum, this study defining the extent and role of coding and non-coding transcription during key stages of germination expands our understanding of transcriptional mechanisms underlying plant developmental transitions.</span></p>',
'date' => '2024-02-26',
'pmid' => 'https://www.nature.com/articles/s41467-024-46082-5',
'doi' => 'https://doi.org/10.1038/s41467-024-46082-5',
'modified' => '2024-02-29 11:56:56',
'created' => '2024-02-29 11:56:56',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 18 => array(
'id' => '4910',
'name' => 'The transcriptional regulatory network modulating human trophoblast stem cells to extravillous trophoblast differentiation',
'authors' => 'Kim M. et al.',
'description' => '<p><span>During human pregnancy, extravillous trophoblasts play crucial roles in placental invasion into the maternal decidua and spiral artery remodeling. However, regulatory factors and their action mechanisms modulating human extravillous trophoblast specification have been unknown. By analyzing dynamic changes in transcriptome and enhancer profile during human trophoblast stem cell to extravillous trophoblast differentiation, we define stage-specific regulators, including an early-stage transcription factor, TFAP2C, and multiple late-stage transcription factors. Loss-of-function studies confirm the requirement of all transcription factors identified for adequate differentiation, and we reveal that the dynamic changes in the levels of TFAP2C are essential. Notably, TFAP2C pre-occupies the regulatory elements of the inactive extravillous trophoblast-active genes during the early stage of differentiation, and the late-stage transcription factors directly activate extravillous trophoblast-active genes, including themselves as differentiation further progresses, suggesting sequential actions of transcription factors assuring differentiation. Our results reveal stage-specific transcription factors and their inter-connected regulatory mechanisms modulating extravillous trophoblast differentiation, providing a framework for understanding early human placentation and placenta-related complications.</span></p>',
'date' => '2024-02-12',
'pmid' => 'https://www.nature.com/articles/s41467-024-45669-2',
'doi' => 'https://doi.org/10.1038/s41467-024-45669-2',
'modified' => '2024-02-15 10:43:52',
'created' => '2024-02-15 10:43:52',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 19 => array(
'id' => '4928',
'name' => 'DMSO derives Trophectoderm and Clonal Blastoid from Single Human Pluripotent Stem Cell',
'authors' => 'Alsolami S. et al.',
'description' => '<p><span>Human naïve pluripotent stem cells (nPSCs) can differentiate into extra-embryonic trophectoderm (TE), a critical step in the generation of the integrated embryo model termed blastoid. The current paradigm of blastoid generation necessitates the aggregation of dozens of nPSCs treated with multiple small molecule inhibitors, growth factors, or genetic modifications to initiate TE differentiation. The presence of complex crosstalk among pathways and cellular heterogeneity in these models complicates mechanistic study and genetic screens. Here, we show that a single small molecule, dimethyl sulfoxide (DMSO), potently induces TE differentiation in basal medium without pharmacological and genetic perturbations. DMSO enhances blastoid generation and, more importantly, is sufficient for blastoid generation by itself. DMSO blastoids resemble blastocysts in morphology and lineage composition. DMSO induces blastoid formation through PKC signaling and cell cycle regulation. Lastly, DMSO enables single nPSC-derived clonal blastoids, which could facilitate genetic screens for mechanistic understanding of human embryogenesis.</span></p>',
'date' => '2024-01-01',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2023.12.31.573770v1.abstract',
'doi' => 'https://doi.org/10.1101/2023.12.31.573770',
'modified' => '2024-03-27 15:18:04',
'created' => '2024-03-27 15:18:04',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 20 => array(
'id' => '4887',
'name' => 'In vitro production of cat-restricted Toxoplasma pre-sexual stages',
'authors' => 'Antunes, A.V. et al.',
'description' => '<p><span>Sexual reproduction of </span><i>Toxoplasma gondii</i><span>, confined to the felid gut, remains largely uncharted owing to ethical concerns regarding the use of cats as model organisms. Chromatin modifiers dictate the developmental fate of the parasite during its multistage life cycle, but their targeting to stage-specific cistromes is poorly described</span><sup><a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 1" title="Farhat, D. C. et al. A MORC-driven transcriptional switch controls Toxoplasma developmental trajectories and sexual commitment. Nat. Microbiol. 5, 570–583 (2020)." href="https://www.nature.com/articles/s41586-023-06821-y#ref-CR1" id="ref-link-section-d277698175e527">1</a>,<a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 2" title="Bougdour, A. et al. Drug inhibition of HDAC3 and epigenetic control of differentiation in Apicomplexa parasites. J. Exp. Med. 206, 953–966 (2009)." href="https://www.nature.com/articles/s41586-023-06821-y#ref-CR2" id="ref-link-section-d277698175e530">2</a></sup><span>. Here we found that the transcription factors AP2XII-1 and AP2XI-2 operate during the tachyzoite stage, a hallmark of acute toxoplasmosis, to silence genes necessary for merozoites, a developmental stage critical for subsequent sexual commitment and transmission to the next host, including humans. Their conditional and simultaneous depletion leads to a marked change in the transcriptional program, promoting a full transition from tachyzoites to merozoites. These in vitro-cultured pre-gametes have unique protein markers and undergo typical asexual endopolygenic division cycles. In tachyzoites, AP2XII-1 and AP2XI-2 bind DNA as heterodimers at merozoite promoters and recruit MORC and HDAC3 (ref. </span><sup><a data-track="click" data-track-action="reference anchor" data-track-label="link" data-test="citation-ref" aria-label="Reference 1" title="Farhat, D. C. et al. A MORC-driven transcriptional switch controls Toxoplasma developmental trajectories and sexual commitment. Nat. Microbiol. 5, 570–583 (2020)." href="https://www.nature.com/articles/s41586-023-06821-y#ref-CR1" id="ref-link-section-d277698175e534">1</a></sup><span>), thereby limiting chromatin accessibility and transcription. Consequently, the commitment to merogony stems from a profound epigenetic rewiring orchestrated by AP2XII-1 and AP2XI-2. Successful production of merozoites in vitro paves the way for future studies on<span> </span></span><i>Toxoplasma</i><span><span> </span>sexual development without the need for cat infections and holds promise for the development of therapies to prevent parasite transmission.</span></p>',
'date' => '2023-12-13',
'pmid' => 'https://www.nature.com/articles/s41586-023-06821-y',
'doi' => 'https://doi.org/10.1038/s41586-023-06821-y',
'modified' => '2023-12-18 10:40:50',
'created' => '2023-12-18 10:40:50',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 21 => array(
'id' => '4939',
'name' => 'Vaginal microbes alter epithelial transcriptomic and epigenomic modifications providing insight into the molecular mechanisms for susceptibility to adverse reproductive outcomes',
'authors' => 'Elovitz M. et al.',
'description' => '<p><span>The cervicovaginal microbiome is highly associated with women's health with microbial communities dominated by </span><i>Lactobacillus</i><span><span> </span>spp. being considered optimal. Conversely, a lack of lactobacilli and a high abundance of strict and facultative anaerobes including<span> </span></span><i>Gardnerella vaginalis</i><span>, have been associated with adverse reproductive outcomes. However, the molecular pathways modulated by microbe interactions with the cervicovaginal epithelia remain unclear. Using RNA-sequencing, we characterize the<span> </span></span><i>in vitro</i><span><span> </span>cervicovaginal epithelial transcriptional response to different vaginal bacteria and their culture supernatants. We showed that<span> </span></span><i>G. vaginalis</i><span><span> </span>upregulated genes were associated with an activated innate immune response including anti-microbial peptides and inflammasome pathways, represented by NLRP3-mediated increases in caspase-1, IL-1β and cell death. Cervicovaginal epithelial cells exposed to<span> </span></span><i>L. crispatus</i><span><span> </span>showed limited transcriptomic changes, while exposure to<span> </span></span><i>L. crispatus</i><span><span> </span>culture supernatants resulted in a shift in the epigenomic landscape of cervical epithelial cells. ATAC-sequencing confirmed epigenetic changes with reduced chromatin accessibility. This study reveals new insight into host-microbe interactions in the lower reproductive tract and suggest potential therapeutic strategies leveraging the vaginal microbiome to improve reproductive health.</span></p>',
'date' => '2023-11-16',
'pmid' => 'https://pubmed.ncbi.nlm.nih.gov/38014044/',
'doi' => '10.21203/rs.3.rs-3580132/v1',
'modified' => '2024-06-07 15:47:48',
'created' => '2024-06-07 15:47:48',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 22 => array(
'id' => '4927',
'name' => 'Inflammatory stress-mediated chromatin changes underlie dysfunction in endothelial cells',
'authors' => 'Liu H. et al.',
'description' => '<p><span>Inflammatory stresses underlie endothelial dysfunction and contribute to the development of chronic cardiovascular disorders such as atherosclerosis and vascular fibrosis. The initial transcriptional response of endothelial cells to pro-inflammatory cytokines such as TNF-alpha is well established. However, very few studies uncover the effects of inflammatory stresses on chromatin architecture. We used integrative analysis of ATAC-seq and RNA-seq data to investigate chromatin alterations in human endothelial cells in response to TNF-alpha and febrile-range heat stress exposure. Multi-omics data analysis suggests a correlation between the transcription of stress-related genes and endothelial dysfunction drivers with chromatin regions exhibiting differential accessibility. Moreover, microscopy identified the dynamics in the nuclear organization, specifically, the changes in a subset of heterochromatic nucleoli-associated chromatin domains, the centromeres. Upon inflammatory stress exposure, the centromeres decreased association with nucleoli in a p38-dependent manner and increased the number of transcripts from pericentromeric regions. Overall, we provide two lines of evidence that suggest chromatin alterations in vascular endothelial cells during inflammatory stresses.</span></p>',
'date' => '2023-10-16',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10614786/',
'doi' => '10.1101/2023.10.11.561959',
'modified' => '2024-03-27 15:15:21',
'created' => '2024-03-27 15:15:21',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 23 => array(
'id' => '4880',
'name' => 'Dynamic PRC1-CBX8 stabilizes a porous structure of chromatin condensates',
'authors' => 'Uckelmann M. et al.',
'description' => '<p><span>The compaction of chromatin is a prevalent paradigm in gene repression. Chromatin compaction is commonly thought to repress transcription by restricting chromatin accessibility. However, the spatial organisation and dynamics of chromatin compacted by gene-repressing factors are unknown. Using cryo-electron tomography, we solved the three dimensional structure of chromatin condensed by the Polycomb Repressive Complex 1 (PRC1) in a complex with CBX8. PRC1-condensed chromatin is porous and stabilised through multivalent dynamic interactions of PRC1 with chromatin. Within condensates, PRC1 remains dynamic while maintaining a static chromatin structure. In differentiated mouse embryonic stem cells, CBX8-bound chromatin remains accessible. These findings challenge the idea of rigidly compacted polycomb domains and instead provides a mechanistic framework for dynamic and accessible PRC1-chromatin condensates.</span></p>',
'date' => '2023-05-09',
'pmid' => 'https://www.biorxiv.org/content/10.1101/2023.05.08.539931v1.abstract',
'doi' => 'https://doi.org/10.1101/2023.05.08.539931',
'modified' => '2023-11-10 15:43:37',
'created' => '2023-11-10 15:43:37',
'ProductsPublication' => array(
[maximum depth reached]
)
),
(int) 24 => array(
'id' => '4810',
'name' => 'UMSBP2 is chromatin remodeler that functions in regulation of geneexpression and suppression of antigenic variation in trypanosomes.',
'authors' => 'Soni A. et al.',
'description' => '<p><span>Universal Minicircle Sequence binding proteins (UMSBPs) are CCHC-type zinc-finger proteins that bind the single-stranded G-rich UMS sequence, conserved at the replication origins of minicircles in the kinetoplast DNA, the mitochondrial genome of kinetoplastids. Trypanosoma brucei UMSBP2 has been recently shown to colocalize with telomeres and to play an essential role in chromosome end protection. Here we report that TbUMSBP2 decondenses in vitro DNA molecules, which were condensed by core histones H2B, H4 or linker histone H1. DNA decondensation is mediated via protein-protein interactions between TbUMSBP2 and these histones, independently of its previously described DNA binding activity. Silencing of the TbUMSBP2 gene resulted in a significant decrease in the disassembly of nucleosomes in T. brucei chromatin, a phenotype that could be reverted, by supplementing the knockdown cells with TbUMSBP2. Transcriptome analysis revealed that silencing of TbUMSBP2 affects the expression of multiple genes in T. brucei, with a most significant effect on the upregulation of the subtelomeric variant surface glycoproteins (VSG) genes, which mediate the antigenic variation in African trypanosomes. These observations suggest that UMSBP2 is a chromatin remodeling protein that functions in the regulation of gene expression and plays a role in the control of antigenic variation in T. brucei.</span></p>',
'date' => '2023-05-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/37207337',
'doi' => '10.1093/nar/gkad402',
'modified' => '2023-06-15 08:54:17',
'created' => '2023-06-13 21:11:31',
'ProductsPublication' => array(
[maximum depth reached]
)
)
),
'Testimonial' => array(),
'Area' => array(),
'SafetySheet' => array(
(int) 0 => array(
'id' => '2440',
'name' => 'ATAC-seq kit SDS US en',
'language' => 'en',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-US-en-1_0.pdf',
'countries' => 'US',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 1 => array(
'id' => '2438',
'name' => 'ATAC-seq kit SDS GB en',
'language' => 'en',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-GB-en-1_0.pdf',
'countries' => 'GB',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 2 => array(
'id' => '2437',
'name' => 'ATAC-seq kit SDS FR fr',
'language' => 'fr',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-FR-fr-1_0.pdf',
'countries' => 'FR',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 3 => array(
'id' => '2433',
'name' => 'ATAC-seq kit SDS BE fr',
'language' => 'fr',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-BE-fr-1_0.pdf',
'countries' => 'BE',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 4 => array(
'id' => '2436',
'name' => 'ATAC-seq kit SDS ES es',
'language' => 'es',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-ES-es-1_0.pdf',
'countries' => 'ES',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 5 => array(
'id' => '2439',
'name' => 'ATAC-seq kit SDS JP ja',
'language' => 'ja',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-JP-ja-1_0.pdf',
'countries' => 'JP',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 6 => array(
'id' => '2435',
'name' => 'ATAC-seq kit SDS DE de',
'language' => 'de',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-DE-de-1_0.pdf',
'countries' => 'DE',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
),
(int) 7 => array(
'id' => '2434',
'name' => 'ATAC-seq kit SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-BE-nl-1_0.pdf',
'countries' => 'BE',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
[maximum depth reached]
)
)
)
)
$meta_canonical = 'https://dev.diagenode.com/en/p/atac-seq-kit-24rxns'
$fromEmail = 'dev.diagenode.com'
$country = 'US'
$countries_allowed = array(
(int) 0 => 'CA',
(int) 1 => 'US',
(int) 2 => 'IE',
(int) 3 => 'GB',
(int) 4 => 'DK',
(int) 5 => 'NO',
(int) 6 => 'SE',
(int) 7 => 'FI',
(int) 8 => 'NL',
(int) 9 => 'BE',
(int) 10 => 'LU',
(int) 11 => 'FR',
(int) 12 => 'DE',
(int) 13 => 'CH',
(int) 14 => 'AT',
(int) 15 => 'ES',
(int) 16 => 'IT',
(int) 17 => 'PT'
)
$outsource = false
$other_formats = array(
(int) 0 => array(
'id' => '3193',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<p><a href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p><strong>ATAC-seq</strong>, Assay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the <strong>transposase Tn5</strong> which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
<p>The Diagenode’s <b>ATAC-</b><b>seq</b><b> kit </b>is based on a highly validated protocol optimized for <b>50,000 </b><b>cells</b><b> per </b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The primer indexes for multiplexing are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell </b><b>requirement</b><b>: </b><b>50,000 </b><b>cells / </b><b>rxn</b></li>
<li><b>Robust protocol </b>with <b>high reproducibility </b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong> and <b>efficient DNA capture </b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids </b><b>over-amplification </b></li>
<li>Allows adaptation/flexibility for <b>more challenging samples </b>to succeed with library prep.</li>
<li>Gives <strong>early indication</strong> if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p><span>Looking for ATAC-seq on tissue? Please, go to: </span><a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack: <a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"> 0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"> (Tn5 transposase) </a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"> Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '8 rxns',
'catalog_number' => 'C01080001',
'old_catalog_number' => '',
'sf_code' => 'C01080001-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '615',
'price_USD' => '615',
'price_GBP' => '535',
'price_JPY' => '110700',
'price_CNY' => '',
'price_AUD' => '1538',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-8rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment (C01080001)| Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2023-05-05 07:57:08',
'created' => '2021-06-24 16:47:47',
'ProductsGroup' => array(
'id' => '397',
'product_id' => '3193',
'group_id' => '349'
)
)
)
$pro = array(
'id' => '3193',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<p><a href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p><strong>ATAC-seq</strong>, Assay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the <strong>transposase Tn5</strong> which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
<p>The Diagenode’s <b>ATAC-</b><b>seq</b><b> kit </b>is based on a highly validated protocol optimized for <b>50,000 </b><b>cells</b><b> per </b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The primer indexes for multiplexing are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell </b><b>requirement</b><b>: </b><b>50,000 </b><b>cells / </b><b>rxn</b></li>
<li><b>Robust protocol </b>with <b>high reproducibility </b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong> and <b>efficient DNA capture </b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids </b><b>over-amplification </b></li>
<li>Allows adaptation/flexibility for <b>more challenging samples </b>to succeed with library prep.</li>
<li>Gives <strong>early indication</strong> if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p><span>Looking for ATAC-seq on tissue? Please, go to: </span><a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack: <a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"> 0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"> (Tn5 transposase) </a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"> Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '8 rxns',
'catalog_number' => 'C01080001',
'old_catalog_number' => '',
'sf_code' => 'C01080001-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '615',
'price_USD' => '615',
'price_GBP' => '535',
'price_JPY' => '110700',
'price_CNY' => '',
'price_AUD' => '1538',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-8rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment (C01080001)| Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2023-05-05 07:57:08',
'created' => '2021-06-24 16:47:47',
'ProductsGroup' => array(
'id' => '397',
'product_id' => '3193',
'group_id' => '349'
)
)
$edit = ''
$testimonials = ''
$featured_testimonials = ''
$related_products = ''
$rrbs_service = array(
(int) 0 => (int) 1894,
(int) 1 => (int) 1895
)
$chipseq_service = array(
(int) 0 => (int) 2683,
(int) 1 => (int) 1835,
(int) 2 => (int) 1836,
(int) 3 => (int) 2684,
(int) 4 => (int) 1838,
(int) 5 => (int) 1839,
(int) 6 => (int) 1856
)
$labelize = object(Closure) {
}
$old_catalog_number = ''
$country_code = 'US'
$other_format = array(
'id' => '3193',
'antibody_id' => null,
'name' => 'ATAC-seq kit',
'description' => '<p><a href="https://www.diagenode.com/files/products/kits/atacseq-kit-manual.pdf"><img src="https://www.diagenode.com/img/buttons/bt-manual.png" /></a></p>
<p><strong>ATAC-seq</strong>, Assay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology for genome-wide mapping of accessible chromatin. The technology is based on the use of the <strong>transposase Tn5</strong> which cuts exposed open chromatin and simultaneously ligates adapters for subsequent amplification and sequencing. ATAC-seq methods allow you to:</p>
<ul>
<li> Gain insight into gene regulation and understand open chromatin signatures</li>
<li> Determine nucleosome positions at single nucleotide resolution</li>
<li> Uncover transcription factor (TF) occupancy</li>
</ul>
<p>The Diagenode’s <b>ATAC-</b><b>seq</b><b> kit </b>is based on a highly validated protocol optimized for <b>50,000 </b><b>cells</b><b> per </b><b>reaction</b>. The kit includes the reagents for cell lysis and nuclei extraction, tagmentation and DNA purification as well as for library amplification. The primer indexes for multiplexing are not included in the kit and must be purchased separately.</p>
<h4><span style="font-weight: 400;">ATAC-seq kit features:</span></h4>
<ul>
<li><b>Cell </b><b>requirement</b><b>: </b><b>50,000 </b><b>cells / </b><b>rxn</b></li>
<li><b>Robust protocol </b>with <b>high reproducibility </b>between replicates and repetitive experiments</li>
<li><strong>Easy</strong> and <b>efficient DNA capture </b>after the tagmentation reaction using Diagenode`s MicroChIP DiaPure columns (included)</li>
<li>Additional qPCR step to determine the number of cycles needed for library amplification: </li>
<ul type="”square”">
<li><b>Avoids </b><b>over-amplification </b></li>
<li>Allows adaptation/flexibility for <b>more challenging samples </b>to succeed with library prep.</li>
<li>Gives <strong>early indication</strong> if the experiment does not work (no qPCR amplification)</li>
</ul>
</ul>
<p><span>Looking for ATAC-seq on tissue? Please, go to: </span><a href="https://www.diagenode.com/en/p/ATAC-seq-package-tissue-C01080006">ATAC-seq package for tissue</a></p>',
'label1' => 'Method overview',
'info1' => '<p><strong>ATAC-seq</strong>, <strong>A</strong>ssay for <strong>T</strong>ransposase-<strong>A</strong>ccessible <strong>C</strong>hromatin, followed by next generation sequencing, is a key technology to easily identify the <strong>open regions of the chromatin.</strong> The protocol consists of <strong>3 steps</strong>: <strong>nuclei preparation</strong>, <strong>tagmentation</strong> and <strong>library amplification</strong>. First, the cells undergo the lysis, ending with the crude nuclei. Then, the nuclei are incubated with a tagmentase (Tn5 transposase), which cuts the genomic regions associated with open chromatin and inserts the sequencing adaptors. Finally, the generated libraries are amplified and can be used for sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only, giving a map of the chromatin status in the whole genome of the sample.</p>
<p><img src="https://www.diagenode.com/img/product/kits/workflow-atac-seq.png" alt="ATAC-seq kit workflow" width="600px" caption="false" /></p>',
'label2' => 'Example of results',
'info2' => '<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig1.png" alt="library prepared with the Diagenode ATAC-seq kit " width="500px" caption="false" /></p>
<p><strong>Figure 1.</strong>Representative Bioanalyzer profile of an ATAC-seq library prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig2.png" alt="Diagenode ATAC-seq kit " caption="false" width="951" height="148" /></p>
<p><strong>Figure 2.</strong> Main ATAC-seq alignment and peak calling statistics of 3 replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells. (Mapping efficiency: Percentage of non-mitochondrial reads that mapped to the reference genome. Uniquely mapped ratio: Proportion of mapped reads that map to only one location on the reference genome (hg19). Peaks: Number of peaks (open chromatin regions) identified by MACS2 for each sample. FRiP - Fraction of reads in peaks: Percentage of reads in peaks, with respect to the number of uniquely mapped reads. Sequencing was realized in paired-end mode 50 base pairs (PE50) on an Illumina NovaSeq6000.)</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3a.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig3b.png" alt="Assay for Transposase-Accessible Chromatin" width="500px" caption="false" /></p>
<p><strong>Figure 3</strong> Sequencing profiles of ATAC-seq library (3 replicates) prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>
<p><img src="https://www.diagenode.com/img/product/kits/atacseq-fig4.png" alt=" open chromatin regions" caption="false" width="383" height="739" /></p>
<p><strong>Figure 4. </strong><br /> Heatmap around TSS of three ATAC-seq replicates prepared with the Diagenode ATAC-seq kit and 24 UDI for tagmented libraries (Cat. No. C01011034) on 50,000 nuclei from K562 cells.</p>',
'label3' => 'Additional solutions for ATAC-seq kit',
'info3' => '<p><a href="https://www.diagenode.com/en/categories/primer-indexes-for-tagmented-libraries">Primer indexes for tagmented libraries</a></p>
<p>Magnetic rack: <a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit">DiaMag</a><a href="https://www.diagenode.com/en/p/diamag02-magnetic-rack-1-unit"> 0.2 ml – Cat. No. B04000001</a></p>
<p>Additional supplies (included in the kit and available separately):</p>
<ul>
<li><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">Tagmentase</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30"> (Tn5 transposase) </a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">loaded</a><a href="https://www.diagenode.com/en/p/tagmentase-loaded-30">, Cat. No. C01070012</a></li>
<li><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x">Tagmentation</a><a href="https://www.diagenode.com/en/p/tagmentation-buffer-2x"> Buffer (2x), Cat. No. C01019043</a></li>
<li><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">MicroChIP</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">DiaPure</a> <a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">columns</a><a href="https://www.diagenode.com/en/p/microchip-diapure-columns-50-rxns">, Cat. No. C03040001</a></li>
</ul>',
'format' => '8 rxns',
'catalog_number' => 'C01080001',
'old_catalog_number' => '',
'sf_code' => 'C01080001-',
'type' => 'RFR',
'search_order' => '',
'price_EUR' => '615',
'price_USD' => '615',
'price_GBP' => '535',
'price_JPY' => '110700',
'price_CNY' => '',
'price_AUD' => '1538',
'country' => 'ALL',
'except_countries' => 'None',
'quote' => false,
'in_stock' => false,
'featured' => false,
'no_promo' => false,
'online' => true,
'master' => false,
'last_datasheet_update' => '',
'slug' => 'atac-seq-kit-8rxns',
'meta_title' => 'ATAC-seq kit for open chromatin assessment (C01080001)| Diagenode ',
'meta_keywords' => '',
'meta_description' => 'Diagenode’s ATAC-seq kit provides a robust protocol for assessing genome-wide chromatin accessibility',
'modified' => '2023-05-05 07:57:08',
'created' => '2021-06-24 16:47:47',
'ProductsGroup' => array(
'id' => '397',
'product_id' => '3193',
'group_id' => '349'
)
)
$label = '<img src="/img/banners/banner-customizer-back.png" alt=""/>'
$document = array(
'id' => '1139',
'name' => 'ATAC-seq Kit - Manual',
'description' => '<p class="p1">Gene expression is carefully regulated in the cells in order to manage a wide range of biological functions. The structure of chromatin is quite dynamic and contributes to this crucial regulatory process.</p>
<p class="p1">ATAC-seq, Assay for Transposase-Accessible Chromatin, followed by nextgeneration sequencing, is a key technology to easily identify the “open” regions of the chromatin, which are usually associated with permissive gene expression. Indeed, the nuclei of the samples are incubated with a transposase, and only the genomic regions associated with open chromatin will be accessible to this transposase. During the process those regions will be cut and sequencing adaptors will be added, allowing their sequencing. High-throughput sequencing will then detect peaks, in open regions of the chromatin only., giving a map of the chromatin status in the whole genome of the sample.</p>
<p class="p1">The Diagenode’s ATAC-seq kit is based on a highly validated protocol, used for years in our Epigenomics Profiling Services offer and takes advantage of many successful Diagenode’s tools, such as the loaded Tagmentase (Tn5 transposase), the MicroChIP DiaPure Columns and the Primer indexes for tagmented libraries kits.</p>',
'image_id' => null,
'type' => 'Manual',
'url' => 'files/products/kits/atacseq-kit-manual.pdf',
'slug' => 'atacseq-kit-manual',
'meta_keywords' => '',
'meta_description' => '',
'modified' => '2021-06-25 09:51:59',
'created' => '2021-06-25 09:51:59',
'ProductsDocument' => array(
'id' => '3130',
'product_id' => '3194',
'document_id' => '1139'
)
)
$sds = array(
'id' => '2434',
'name' => 'ATAC-seq kit SDS BE nl',
'language' => 'nl',
'url' => 'files/SDS/ATAC-seq/SDS-C01080001_C01080002-ATAC-seq_kit-BE-nl-1_0.pdf',
'countries' => 'BE',
'modified' => '2022-08-30 10:36:01',
'created' => '2022-08-30 10:36:01',
'ProductsSafetySheet' => array(
'id' => '4140',
'product_id' => '3194',
'safety_sheet_id' => '2434'
)
)
$publication = array(
'id' => '4810',
'name' => 'UMSBP2 is chromatin remodeler that functions in regulation of geneexpression and suppression of antigenic variation in trypanosomes.',
'authors' => 'Soni A. et al.',
'description' => '<p><span>Universal Minicircle Sequence binding proteins (UMSBPs) are CCHC-type zinc-finger proteins that bind the single-stranded G-rich UMS sequence, conserved at the replication origins of minicircles in the kinetoplast DNA, the mitochondrial genome of kinetoplastids. Trypanosoma brucei UMSBP2 has been recently shown to colocalize with telomeres and to play an essential role in chromosome end protection. Here we report that TbUMSBP2 decondenses in vitro DNA molecules, which were condensed by core histones H2B, H4 or linker histone H1. DNA decondensation is mediated via protein-protein interactions between TbUMSBP2 and these histones, independently of its previously described DNA binding activity. Silencing of the TbUMSBP2 gene resulted in a significant decrease in the disassembly of nucleosomes in T. brucei chromatin, a phenotype that could be reverted, by supplementing the knockdown cells with TbUMSBP2. Transcriptome analysis revealed that silencing of TbUMSBP2 affects the expression of multiple genes in T. brucei, with a most significant effect on the upregulation of the subtelomeric variant surface glycoproteins (VSG) genes, which mediate the antigenic variation in African trypanosomes. These observations suggest that UMSBP2 is a chromatin remodeling protein that functions in the regulation of gene expression and plays a role in the control of antigenic variation in T. brucei.</span></p>',
'date' => '2023-05-01',
'pmid' => 'https://www.ncbi.nlm.nih.gov/pubmed/37207337',
'doi' => '10.1093/nar/gkad402',
'modified' => '2023-06-15 08:54:17',
'created' => '2023-06-13 21:11:31',
'ProductsPublication' => array(
'id' => '6971',
'product_id' => '3194',
'publication_id' => '4810'
)
)
$externalLink = ' <a href="https://www.ncbi.nlm.nih.gov/pubmed/37207337" target="_blank"><i class="fa fa-external-link"></i></a>'CustomErrorHandler::handleError() - APP/Lib/CustomErrorHandler.php, line 9
include - APP/View/Products/view.ctp, line 755
View::_evaluate() - CORE/Cake/View/View.php, line 971
View::_render() - CORE/Cake/View/View.php, line 933
View::render() - CORE/Cake/View/View.php, line 473
Controller::render() - CORE/Cake/Controller/Controller.php, line 963
ProductsController::slug() - APP/Controller/ProductsController.php, line 1055
ReflectionMethod::invokeArgs() - [internal], line ??
Controller::invokeAction() - CORE/Cake/Controller/Controller.php, line 491
Dispatcher::_invoke() - CORE/Cake/Routing/Dispatcher.php, line 193
Dispatcher::dispatch() - CORE/Cake/Routing/Dispatcher.php, line 167
[main] - APP/webroot/index.php, line 118
×